This is a preprint.
Evolvability of cancer-associated genes under APOBEC3A/B selection
- PMID: 38106028
- PMCID: PMC10723265
- DOI: 10.1101/2023.08.27.554991
Evolvability of cancer-associated genes under APOBEC3A/B selection
Update in
-
Evolvability of cancer-associated genes under APOBEC3A/B selection.iScience. 2024 Mar 6;27(4):109433. doi: 10.1016/j.isci.2024.109433. eCollection 2024 Apr 19. iScience. 2024. PMID: 38550998 Free PMC article.
Abstract
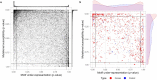
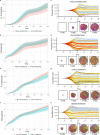
Evolvability is an emergent hallmark of cancer that depends on intra-tumor heterogeneity and, ultimately, genetic variation. Mutations generated by APOBEC3 cytidine deaminases can contribute to genetic variation and the consequences of APOBEC activation differ depending on the stage of cancer, with the most significant impact observed during the early stages. However, how APOBEC activity shapes evolutionary patterns of genes in the host genome and differential impacts on cancer-associated and non-cancer genes remain unclear. Analyzing over 40,000 human protein-coding transcripts, we identified distinct distribution patterns of APOBEC3A/B TC motifs between cancer-related genes and controls, suggesting unique associations with cancer. Studying a bat species with many more APOBEC3 genes, we found diverse motif patterns in orthologs of cancer genes compared to controls, similar to humans and suggesting APOBEC evolution to reduce impacts on the genome rather than the converse. Simulations confirmed that APOBEC-induced heterogeneity enhances cancer evolution, shaping clonal dynamics through bimodal introduction of mutations in certain classes of genes. Our results suggest that a major consequence of the bimodal distribution of APOBEC affects greater cancer heterogeneity.
Keywords: APOBEC; Evolvability; Intra-tumor heterogeneity; Neutral evolution.
Conflict of interest statement
Declaration of interests The authors declare no competing interest.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
