This is a preprint.
Regulation of transcription patterns, poly-ADP-ribose, and RNA-DNA hybrids by the ATM protein kinase
- PMID: 38106035
- PMCID: PMC10723464
- DOI: 10.1101/2023.12.06.570417
Regulation of transcription patterns, poly-ADP-ribose, and RNA-DNA hybrids by the ATM protein kinase
Update in
-
Regulation of transcription patterns, poly(ADP-ribose), and RNA-DNA hybrids by the ATM protein kinase.Cell Rep. 2024 Mar 26;43(3):113896. doi: 10.1016/j.celrep.2024.113896. Epub 2024 Mar 4. Cell Rep. 2024. PMID: 38442018 Free PMC article.
Abstract
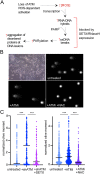
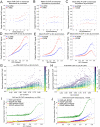
The ATM protein kinase is a master regulator of the DNA damage response and also an important sensor of oxidative stress. Analysis of gene expression in Ataxia-telangiectasia patient brain tissue shows that large-scale transcriptional changes occur in patient cerebellum that correlate with expression level and GC content of transcribed genes. In human neuron-like cells in culture we map locations of poly-ADP-ribose and RNA-DNA hybrid accumulation genome-wide with ATM inhibition and find that these marks also coincide with high transcription levels, active transcription histone marks, and high GC content. Antioxidant treatment reverses the accumulation of R-loops in transcribed regions, consistent with the central role of ROS in promoting these lesions. Based on these results we postulate that transcription-associated lesions accumulate in ATM-deficient cells and that the single-strand breaks and PARylation at these sites ultimately generate changes in transcription that compromise cerebellum function and lead to neurodegeneration over time in A-T patients.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures




References
-
- Barzilai A., Rotman G., and Shiloh Y. (2002). ATM deficiency and oxidative stress: a new dimension of defective response to DNA damage. DNA Repair 1, 3–25. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
