This is a preprint.
Dystrophin deficiency impairs cell junction formation during embryonic myogenesis
- PMID: 38106055
- PMCID: PMC10723310
- DOI: 10.1101/2023.12.05.569919
Dystrophin deficiency impairs cell junction formation during embryonic myogenesis
Update in
-
Dystrophin deficiency impairs cell junction formation during embryonic myogenesis from pluripotent stem cells.iScience. 2024 Jun 11;27(7):110242. doi: 10.1016/j.isci.2024.110242. eCollection 2024 Jul 19. iScience. 2024. PMID: 39040067 Free PMC article.
Abstract
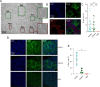
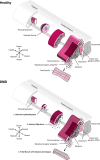
Mutations in the DMD gene lead to Duchenne muscular dystrophy, a severe X-linked neuromuscular disorder that manifests itself as young boys acquire motor functions. DMD is typically diagnosed at 2 to 4 years of age, but the absence of dystrophin negatively impacts muscle structure and function before overt symptoms appear in patients, which poses a serious challenge in the optimization of standards of care. In this report, we investigated the early consequences of dystrophin deficiency during skeletal muscle development. We used single-cell transcriptome profiling to characterize the myogenic trajectory of human pluripotent stem cells and showed that DMD cells bifurcate to an alternative branch when they reach the somite stage. Here, dystrophin deficiency was linked to marked dysregulations of cell junction protein families involved in the cell state transitions characteristic of embryonic somitogenesis. Altogether, this work demonstrates that in vitro, dystrophin deficiency has deleterious effects on cell-cell communication during myogenic development, which should be considered in future therapeutic strategies for DMD.
Keywords: DMD; Duchenne muscular dystrophy; cell junctions; hiPSCs; myogenesis; somite.
Conflict of interest statement
Declaration of interests The authors declare no competing interests
Figures




References
-
- Bushby K., Finkel R., Birnkrant D.J., Case L.E., Clemens P.R., Cripe L., Kaul A., Kinnett K., McDonald C., Pandya S., et al. (2010). Diagnosis and management of Duchenne muscular dystrophy, part 1: diagnosis, and pharmacological and psychosocial management. Lancet Neurol 9, 77–93. 10.1016/S1474-4422(09)70271-6. - DOI - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
