This is a preprint.
Somatic mutation as an explanation for epigenetic aging
- PMID: 38106096
- PMCID: PMC10723383
- DOI: 10.1101/2023.12.08.569638
Somatic mutation as an explanation for epigenetic aging
Update in
-
Somatic mutation as an explanation for epigenetic aging.Nat Aging. 2025 Apr;5(4):709-719. doi: 10.1038/s43587-024-00794-x. Epub 2025 Jan 13. Nat Aging. 2025. PMID: 39806003
Abstract
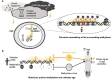
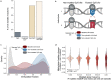
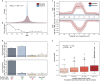
DNA methylation marks have recently been used to build models known as "epigenetic clocks" which predict calendar age. As methylation of cytosine promotes C-to-T mutations, we hypothesized that the methylation changes observed with age should reflect the accrual of somatic mutations, and the two should yield analogous aging estimates. In analysis of multimodal data from 9,331 human individuals, we find that CpG mutations indeed coincide with changes in methylation, not only at the mutated site but also with pervasive remodeling of the methylome out to ±10 kilobases. This one-to-many mapping enables mutation-based predictions of age that agree with epigenetic clocks, including which individuals are aging faster or slower than expected. Moreover, genomic loci where mutations accumulate with age also tend to have methylation patterns that are especially predictive of age. These results suggest a close coupling between the accumulation of sporadic somatic mutations and the widespread changes in methylation observed over the course of life.
Conflict of interest statement
Declaration of Interests T.I. is a cofounder of Serinus and Data4Cure, is on their Scientific Advisory Boards, and has equity interest in both companies. T.I. is on the Scientific Advisory Board of Ideaya BioSciences and has an equity interest. The terms of these arrangements have been reviewed and approved by the University of California San Diego, in accordance with its conflict of interest policies.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
