This is a preprint.
Association of ESR1 germline variants with TP53 somatic variants in breast tumors in a genome-wide study
- PMID: 38106140
- PMCID: PMC10723566
- DOI: 10.1101/2023.12.06.23299442
Association of ESR1 germline variants with TP53 somatic variants in breast tumors in a genome-wide study
Update in
-
Association of ESR1 Germline Variants with TP53 Somatic Variants in Breast Tumors in a Genome-wide Study.Cancer Res Commun. 2024 Jun 27;4(6):1597-1608. doi: 10.1158/2767-9764.CRC-24-0026. Cancer Res Commun. 2024. PMID: 38836758 Free PMC article.
Abstract
Background: In breast tumors, somatic mutation frequencies in TP53 and PIK3CA vary by tumor subtype and ancestry. HER2 positive and triple negative breast cancers (TNBC) have a higher frequency of TP53 somatic mutations than other subtypes. PIK3CA mutations are more frequently observed in hormone receptor positive tumors. Emerging data suggest tumor mutation status is associated with germline variants and genetic ancestry. We aimed to identify germline variants that are associated with somatic TP53 or PIK3CA mutation status in breast tumors.
Methods: A genome-wide association study was conducted using breast cancer mutation status of TP53 and PIK3CA and functional mutation categories including TP53 gain of function (GOF) and loss of function mutations and PIK3CA activating/hotspot mutations. The discovery analysis consisted of 2850 European ancestry women from three datasets. Germline variants showing evidence of association with somatic mutations were selected for validation analyses based on predicted function, allele frequency, and proximity to known cancer genes or risk loci. Candidate variants were assessed for association with mutation status in a multi-ancestry validation study, a Malaysian study, and a study of African American/Black women with TNBC.
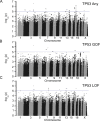
Results: The discovery Germline x Mutation (GxM) association study found five variants associated with one or more TP53 phenotypes with P values <1×10-6, 33 variants associated with one or more TP53 phenotypes with P values <1×10-5, and 44 variants associated with one or more PIK3CA phenotypes with P values <1×10-5. In the multi-ancestry and Malaysian validation studies, germline ESR1 locus variant, rs9383938, was associated with the presence of TP53 mutations overall (P values 6.8×10-5 and 9.8×10-8, respectively) and TP53 GOF mutations (P value 8.4×10-6). Multiple variants showed suggestive evidence of association with PIK3CA mutation status in the validation studies, but none were significant after correction for multiple comparisons.
Conclusions: We found evidence that germline variants were associated with TP53 and PIK3CA mutation status in breast cancers. Variants near the estrogen receptor alpha gene, ESR1, were significantly associated with overall TP53 mutations and GOF mutations. Larger multi-ancestry studies are needed to confirm these findings and determine if these variants contribute to ancestry-specific differences in mutation frequency.
Keywords: ESR1; GWAS; PIK3CA; TP53; breast cancer; germline; germline by somatic interaction; somatic.
Conflict of interest statement
CONFLICTS OF INTEREST HH is on the scientific advisory board for Invitae Genetics, Promega, and Genome Medical and has stock/stock options in Genome Medical and GI OnDemand. None of these are direct conflicts with this study.
Figures

References
-
- Keenan T, Moy B, Mroz EA, Ross K, Niemierko A, Rocco JW et al. Comparison of the genomic landscape between primary breast cancer in African American versus white women and the association of racial differences with tumor recurrence. J Clin Oncol. 2015; 33(31):3621–3627. doi: 10.1200/JCO.2015.62.2126. - DOI - PMC - PubMed
Publication types
Grants and funding
- U24 CA274159/CA/NCI NIH HHS/United States
- U24 CA232979/CA/NCI NIH HHS/United States
- R01 CA184585/CA/NCI NIH HHS/United States
- R01 CA228198/CA/NCI NIH HHS/United States
- R01 MD013452/MD/NIMHD NIH HHS/United States
- P30 CA068485/CA/NCI NIH HHS/United States
- R01 CA202981/CA/NCI NIH HHS/United States
- R01 CA215151/CA/NCI NIH HHS/United States
- P30 CA016058/CA/NCI NIH HHS/United States
- R01 CA228156/CA/NCI NIH HHS/United States
- P30 CA033572/CA/NCI NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- R01 CA255242/CA/NCI NIH HHS/United States
- K24 CA169004/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
