This is a preprint.
p14ARF forms meso-scale assemblies upon phase separation with NPM1
- PMID: 38106181
- PMCID: PMC10723529
- DOI: 10.21203/rs.3.rs-3592059/v1
p14ARF forms meso-scale assemblies upon phase separation with NPM1
Update in
-
p14ARF forms meso-scale assemblies upon phase separation with NPM1.Nat Commun. 2024 Nov 11;15(1):9531. doi: 10.1038/s41467-024-53904-z. Nat Commun. 2024. PMID: 39528457 Free PMC article.
Abstract
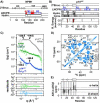
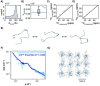
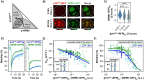
NPM1 is an abundant nucleolar chaperone that, in addition to facilitating ribosome biogenesis, contributes to nucleolar stress responses and tumor suppression through its regulation of the p14 Alternative Reading Frame tumor suppressor protein (p14ARF). Oncogenic stress induces p14ARF to inhibit MDM2, stabilize p53 and arrest the cell cycle. Under non-stress conditions, NPM1 stabilizes p14ARF in nucleoli, preventing its degradation and blocking p53 activation. However, the mechanisms underlying the regulation of p14ARF by NPM1 are unclear because the structural features of the p14ARF-NPM1 complex remain elusive. Here we show that NPM1 sequesters p14ARF within phase-separated condensates, facilitating the assembly of p14ARF into a gel-like meso-scale network. This assembly is mediated by intermolecular contacts formed by hydrophobic residues in an α-helix and β-strands within a partially folded N-terminal domain of p14ARF. Those hydrophobic interactions promote phase separation with NPM1, enhance nucleolar partitioning of p14ARF, restrict p14ARF and NPM1 diffusion within condensates and in nucleoli, and reduce cell viability. Our structural model provides novel insights into the multifaceted chaperone function of NPM1 in nucleoli by mechanistically linking the nucleolar localization of p14ARF to its partial folding and meso-scale assembly upon phase separation with NPM1.
Conflict of interest statement
Additional Declarations: There is NO Competing Interest.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

