Association between gut microbiota and NAFLD/NASH: a bidirectional two-sample Mendelian randomization study
- PMID: 38106475
- PMCID: PMC10722258
- DOI: 10.3389/fcimb.2023.1294826
Association between gut microbiota and NAFLD/NASH: a bidirectional two-sample Mendelian randomization study
Abstract
Background: Recent studies have suggested a relationship between gut microbiota and non-alcoholic fatty liver disease (NAFLD)/nonalcoholic steatohepatitis (NASH). However, the nature and direction of this potential causal relationship are still unclear. This study used two-sample Mendelian randomization (MR) to clarify the potential causal links.
Methods: Summary-level Genome-Wide Association Studies (GWAS) statistical data for gut microbiota and NAFLD/NASH were obtained from MiBioGen and FinnGen respectively. The MR analyses were performed mainly using the inverse-variance weighted (IVW) method, with sensitivity analyses conducted to verify the robustness. Additionally, reverse MR analyses were performed to examine any potential reverse causal associations.
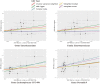
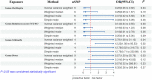
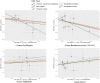
Results: Our analysis, primarily based on the IVW method, strongly supports the existence of causal relationships between four microbial taxa and NAFLD, and four taxa with NASH. Specifically, associations were observed between Enterobacteriales (P =0.04), Enterobacteriaceae (P =0.04), Lachnospiraceae UCG-004 (P =0.02), and Prevotella9 (P =0.04) and increased risk of NAFLD. Dorea (P =0.03) and Veillonella (P =0.04) could increase the risks of NASH while Oscillospira (P =0.04) and Ruminococcaceae UCG-013 (P=0.005) could decrease them. We also identified that NAFLD was found to potentially cause an increased abundance in Holdemania (P =0.007) and Ruminococcus2 (P =0.002). However, we found no evidence of reverse causation in the microbial taxa associations with NASH.
Conclusion: This study identified several specific gut microbiota that are causally related to NAFLD and NASH. Observations herein may provide promising theoretical groundwork for potential prevention and treatment strategies for NAFLD and its progression to NASH in future.
Keywords: Mendelian randomization; causality; gut microbiota; non-alcoholic fatty liver disease; non-alcoholic steatohepatitis.
Copyright © 2023 Zhai, Wu, Zheng, Zhong, Du, Yuan, Peng, Cai and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Metabolites mediate the causal associations between gut microbiota and NAFLD: a Mendelian randomization study.BMC Gastroenterol. 2024 Jul 31;24(1):244. doi: 10.1186/s12876-024-03277-w. BMC Gastroenterol. 2024. PMID: 39085775 Free PMC article.
-
Causal associations between human gut microbiota and cholelithiasis: a mendelian randomization study.Front Cell Infect Microbiol. 2023 May 25;13:1169119. doi: 10.3389/fcimb.2023.1169119. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37305422 Free PMC article.
-
Mendelian randomization suggests a causal relationship between gut microbiota and nonalcoholic fatty liver disease in humans.Medicine (Baltimore). 2024 Mar 22;103(12):e37478. doi: 10.1097/MD.0000000000037478. Medicine (Baltimore). 2024. PMID: 38518048 Free PMC article.
-
Genetic liability of gut microbiota for idiopathic pulmonary fibrosis and lung function: a two-sample Mendelian randomization study.Front Cell Infect Microbiol. 2024 May 22;14:1348685. doi: 10.3389/fcimb.2024.1348685. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38841114 Free PMC article.
-
Causal relationship between gut microbiota and polycystic ovary syndrome: a literature review and Mendelian randomization study.Front Endocrinol (Lausanne). 2024 Feb 1;15:1280983. doi: 10.3389/fendo.2024.1280983. eCollection 2024. Front Endocrinol (Lausanne). 2024. PMID: 38362275 Free PMC article. Review.
Cited by
-
Metabolites mediate the causal associations between gut microbiota and NAFLD: a Mendelian randomization study.BMC Gastroenterol. 2024 Jul 31;24(1):244. doi: 10.1186/s12876-024-03277-w. BMC Gastroenterol. 2024. PMID: 39085775 Free PMC article.
-
The newly proposed dietary index for gut microbiota and its association with the risk of nonalcoholic fatty liver disease: the mediating role of body mass index.Diabetol Metab Syndr. 2025 Jun 21;17(1):237. doi: 10.1186/s13098-025-01801-w. Diabetol Metab Syndr. 2025. PMID: 40544245 Free PMC article.
-
Multifaceted Interactions Between Bile Acids, Their Receptors, and MASH: From Molecular Mechanisms to Clinical Therapeutics.Molecules. 2025 Jul 22;30(15):3066. doi: 10.3390/molecules30153066. Molecules. 2025. PMID: 40807240 Free PMC article. Review.
-
Identification of m6A-associated autophagy genes in non-alcoholic fatty liver.PeerJ. 2024 Feb 29;12:e17011. doi: 10.7717/peerj.17011. eCollection 2024. PeerJ. 2024. PMID: 38436022 Free PMC article.
-
Advances in the acting mechanism and treatment of gut microbiota in metabolic dysfunction-associated steatotic liver disease.Gut Microbes. 2025 Dec;17(1):2500099. doi: 10.1080/19490976.2025.2500099. Epub 2025 May 20. Gut Microbes. 2025. PMID: 40394806 Free PMC article. Review.
References
-
- Adams L. A., Wang Z., Liddle C., Melton P. E., Ariff A., Chandraratna H., et al. . (2020). Bile acids associate with specific gut microbiota, low-level alcohol consumption and liver fibrosis in patients with non-alcoholic fatty liver disease. Liver Int. 40 (6), 1356–1365. doi: 10.1111/liv.14453 - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

