Direct image to subtype prediction for brain tumors using deep learning
- PMID: 38106649
- PMCID: PMC10724115
- DOI: 10.1093/noajnl/vdad139
Direct image to subtype prediction for brain tumors using deep learning
Abstract
Background: Deep Learning (DL) can predict molecular alterations of solid tumors directly from routine histopathology slides. Since the 2021 update of the World Health Organization (WHO) diagnostic criteria, the classification of brain tumors integrates both histopathological and molecular information. We hypothesize that DL can predict molecular alterations as well as WHO subtyping of brain tumors from hematoxylin and eosin-stained histopathology slides.
Methods: We used weakly supervised DL and applied it to three large cohorts of brain tumor samples, comprising N = 2845 patients.
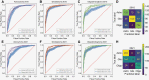
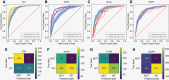
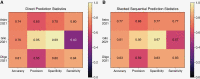
Results: We found that the key molecular alterations for subtyping, IDH and ATRX, as well as 1p19q codeletion, were predictable from histology with an area under the receiver operating characteristic curve (AUROC) of 0.95, 0.90, and 0.80 in the training cohort, respectively. These findings were upheld in external validation cohorts with AUROCs of 0.90, 0.79, and 0.87 for prediction of IDH, ATRX, and 1p19q codeletion, respectively.
Conclusions: In the future, such DL-based implementations could ease diagnostic workflows, particularly for situations in which advanced molecular testing is not readily available.
Keywords: IDH; adult-type diffuse gliomas; deep learning; molecular signatures; subtype.
© The Author(s) 2023. Published by Oxford University Press, the Society for Neuro-Oncology and the European Association of Neuro-Oncology.
Conflict of interest statement
J.N.K. declares consulting services for Owkin, France; DoMore Diagnostics, Norway and Panakeia, UK; furthermore, he holds shares in StratifAI GmbH and has received honoraria for lectures by AstraZeneca, Bayer, Eisai, MSD, BMS, Roche, Pfizer, and Fresenius. No other potential conflict of interest are noted by any of the authors.
Figures

References
-
- WHO Classification of Tumours Editorial Board. Lyon (France): International Agency for Research on Cancer. WHO Classification of Tumours Editorial Board. Central nervous system tumours. [cited 2022 Aug 15] 5th ed., Vol. 7. WHO classification of tumours series; 2021.
LinkOut - more resources
Full Text Sources
