Genome-wide association analysis of time to heading and maturity in bread wheat using 55K microarrays
- PMID: 38107003
- PMCID: PMC10722194
- DOI: 10.3389/fpls.2023.1296197
Genome-wide association analysis of time to heading and maturity in bread wheat using 55K microarrays
Abstract
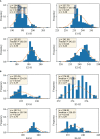
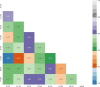
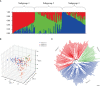
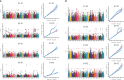
To investigate the genetic mechanisms underlying the reproductive traits (time to flowering and maturity) in wheat and identify candidate genes associated, a phenotypic analysis was conducted on 239 wheat accessions (lines) from around the world. A genome-wide association study (GWAS) of wheat heading and maturity phases was performed using the MLM (Q+K) model in the TASSLE software, combined with the Wheat 55K SNP array. The results revealed significant phenotypic variation in heading and maturity among the wheat accessions across different years, with coefficients of variation ranging from 0.96% to 1.97%. The phenotypic data from different years exhibited excellent correlation, with a genome-wide linkage disequilibrium (LD) attenuation distance of 3 Mb. Population structure analysis, evolutionary tree analysis, and principal component analysis indicated that the 239 wheat accessions formed a relatively homogeneous natural population, which could be divided into three subgroups. The GWAS results identified a total of 293 SNP marker loci that were significantly associated with wheat heading and maturity stages (P ≤ 0.001) in different environments. Among them, nine stable SNP marker loci were consistently detected in multiple environments. These marker loci were distributed on wheat chromosomes 1A、1B、2D、3A、5B、6D and 7A. Each individual locus explained 4.03%-16.06% of the phenotypic variation. Furthermore, through careful analysis of the associated loci with large phenotypic effect values and stable inheritance, a total of nine candidate genes related to wheat heading and maturity stages were identified. These findings have implications for molecular marker-assisted selection breeding programs targeting specific wheat traits at the heading and maturity stages. In summary, this study conducted a comprehensive GWAS of wheat heading and maturity phases, revealing significant associations between genetic markers and key developmental stages in wheat. The identification of candidate genes and marker loci provides valuable information for further studies on wheat breeding and genetic improvement targeted at enhancing heading and maturity traits.
Keywords: GWAS; candidate genes; heading; maturity; wheat.
Copyright © 2023 Ding, Fang, Gao, Fan, Shi, Yu, Ding, Huang, Wang and Song.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Performing whole-genome association analysis of winter wheat plant height using the 55K chip.Front Plant Sci. 2025 Jan 17;15:1471636. doi: 10.3389/fpls.2024.1471636. eCollection 2024. Front Plant Sci. 2025. PMID: 39898269 Free PMC article.
-
Genome-wide association analysis of wheat stem traits using 55K microarrays.Front Plant Sci. 2025 Jul 24;16:1635721. doi: 10.3389/fpls.2025.1635721. eCollection 2025. Front Plant Sci. 2025. PMID: 40777061 Free PMC article.
-
Identification of favorable SNP alleles and candidate genes for traits related to early maturity via GWAS in upland cotton.BMC Genomics. 2016 Aug 30;17(1):687. doi: 10.1186/s12864-016-2875-z. BMC Genomics. 2016. PMID: 27576450 Free PMC article.
-
Genome-wide association study of heading and flowering dates and construction of its prediction equation in Chinese common wheat.Theor Appl Genet. 2018 Nov;131(11):2271-2285. doi: 10.1007/s00122-018-3181-8. Epub 2018 Sep 14. Theor Appl Genet. 2018. PMID: 30218294 Review.
-
Unraveling the Secrets of Early-Maturity and Short-Duration Bread Wheat in Unpredictable Environments.Plants (Basel). 2024 Oct 12;13(20):2855. doi: 10.3390/plants13202855. Plants (Basel). 2024. PMID: 39458802 Free PMC article. Review.
Cited by
-
Performing whole-genome association analysis of winter wheat plant height using the 55K chip.Front Plant Sci. 2025 Jan 17;15:1471636. doi: 10.3389/fpls.2024.1471636. eCollection 2024. Front Plant Sci. 2025. PMID: 39898269 Free PMC article.
-
Unraveling the genetic basis of heat tolerance and yield in bread wheat: QTN discovery and Its KASP-assisted validation.BMC Plant Biol. 2025 Mar 1;25(1):268. doi: 10.1186/s12870-025-06285-4. BMC Plant Biol. 2025. PMID: 40021958 Free PMC article.
-
Deciphering the genetic basis of novel traits that discriminate useful and non-useful biomass to enhance harvest index in wheat.Plant Genome. 2024 Dec;17(4):e20512. doi: 10.1002/tpg2.20512. Epub 2024 Sep 18. Plant Genome. 2024. PMID: 39295194 Free PMC article.
-
Spatial patterns and key driving factors of wheat harvest index under irrigation and rainfed conditions in arid regions.Front Plant Sci. 2025 Jun 9;16:1614204. doi: 10.3389/fpls.2025.1614204. eCollection 2025. Front Plant Sci. 2025. PMID: 40551775 Free PMC article.
-
Quantitative Trait Loci Mapping of Heading Date in Wheat under Phosphorus Stress Conditions.Genes (Basel). 2024 Aug 31;15(9):1150. doi: 10.3390/genes15091150. Genes (Basel). 2024. PMID: 39336741 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

