A covalent fragment-based strategy targeting a novel cysteine to inhibit activity of mutant EGFR kinase
- PMID: 38107172
- PMCID: PMC10718517
- DOI: 10.1039/d3md00439b
A covalent fragment-based strategy targeting a novel cysteine to inhibit activity of mutant EGFR kinase
Abstract
Several generations of ATP-competitive anti-cancer drugs that inhibit the activity of the intracellular kinase domain of the epidermal growth factor receptor (EGFR) have been developed over the past twenty years. The first-generation of drugs such as gefitinib bind reversibly and were followed by a second-generation such as dacomitinib that harbor an acrylamide moiety that forms a covalent bond with C797 in the ATP binding pocket. Resistance emerges through mutation of the T790 gatekeeper residue to methionine, which introduces steric hindrance to drug binding and increases the Km for ATP. A third generation of drugs, such as osimertinib were developed which were effective against T790M EGFR in which an acrylamide moiety forms a covalent bond with C797, although resistance has emerged by mutation to S797. A fragment-based screen to identify new starting points for an EGFR inhibitor serendipitously identified a fragment that reacted with C775, a previously unexploited residue in the ATP binding pocket for a covalent inhibitor to target. A number of acrylamide containing fragments were identified that selectively reacted with C775. One of these acrylamides was optimized to a highly selective inhibitor with sub-1 μM activity, that is active against T790M, C797S mutant EGFR independent of ATP concentration, providing a potential new strategy for pan-EGFR mutant inhibition.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There is no conflict of interest to declare.
Figures



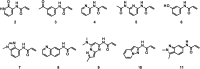

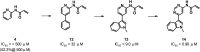


References
LinkOut - more resources
Full Text Sources
Chemical Information
Research Materials
Miscellaneous

