Tsc2 coordinates neuroprogenitor differentiation
- PMID: 38107199
- PMCID: PMC10724693
- DOI: 10.1016/j.isci.2023.108442
Tsc2 coordinates neuroprogenitor differentiation
Abstract
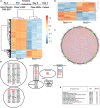
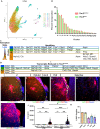
Neural stem cells (NSCs) of the ventricular-subventricular zone (V-SVZ) generate numerous cell types. The uncoupling of mRNA transcript availability and translation occurs during the progression from stem to differentiated states. The mTORC1 kinase pathway acutely controls proteins that regulate mRNA translation. Inhibiting mTORC1 during differentiation is hypothesized to be critical for brain development since somatic mutations of mTORC1 regulators perturb brain architecture. Inactivating mutations of TSC1 or TSC2 genes cause tuberous sclerosis complex (TSC). TSC patients have growths near the striatum and ventricles. Here, it is demonstrated that V-SVZ NSC Tsc2 inactivation causes striatal hamartomas. Tsc2 removal altered translation factors, translatomes, and translational efficiency. Single nuclei RNA sequencing following in vivo loss of Tsc2 revealed changes in NSC activation states. The inability to decouple mRNA transcript availability and translation delayed differentiation leading to the retention of immature phenotypes in hamartomas. Taken together, Tsc2 is required for translational repression and differentiation.
Keywords: Cell biology; Molecular biology; Molecular mechanism of gene regulation; Neuroscience; Stem cells research; Transcriptomics.
© 2023 The Author(s).
Conflict of interest statement
The authors declare that the researchers have no competing financial interests.
Figures



References
-
- Beckervordersandforth R., Tripathi P., Ninkovic J., Bayam E., Lepier A., Stempfhuber B., Kirchhoff F., Hirrlinger J., Haslinger A., Lie D.C., et al. In vivo fate mapping and expression analysis reveals molecular hallmarks of prospectively isolated adult neural stem cells. Cell Stem Cell. 2010;7:744–758. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

