Opposing, spatially-determined epigenetic forces impose restrictions on stochastic olfactory receptor choice
- PMID: 38108811
- PMCID: PMC10727497
- DOI: 10.7554/eLife.87445
Opposing, spatially-determined epigenetic forces impose restrictions on stochastic olfactory receptor choice
Abstract
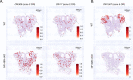
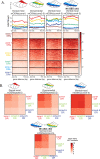
Olfactory receptor (OR) choice represents an example of genetically hardwired stochasticity, where every olfactory neuron expresses one out of ~2000 OR alleles in the mouse genome in a probabilistic, yet stereotypic fashion. Here, we propose that topographic restrictions in OR expression are established in neuronal progenitors by two opposing forces: polygenic transcription and genomic silencing, both of which are influenced by dorsoventral gradients of transcription factors NFIA, B, and X. Polygenic transcription of OR genes may define spatially constrained OR repertoires, among which one OR allele is selected for singular expression later in development. Heterochromatin assembly and genomic compartmentalization of OR alleles also vary across the axes of the olfactory epithelium and may preferentially eliminate ectopically expressed ORs with more dorsal expression destinations from this 'privileged' repertoire. Our experiments identify early transcription as a potential 'epigenetic' contributor to future developmental patterning and reveal how two spatially responsive probabilistic processes may act in concert to establish deterministic, precise, and reproducible territories of stochastic gene expression.
Keywords: chromatin; developmental biology; genomics; heterochromatin; mouse; nuclear architecture; olfaction; patterning.
© 2023, Bashkirova et al.
Conflict of interest statement
EB, NK, KM, CC, JO, LT, IS, AP, BS, GB, XX, IA, RG, AF, SL No competing interests declared, BS is an employee of Prevail Therapeutics, a wholly-owned subsidiary of Eli Lilly and Company, BM Reviewing editor, eLife
Figures












Update of
-
Opposing, spatially-determined epigenetic forces impose restrictions on stochastic olfactory receptor choice.bioRxiv [Preprint]. 2023 Sep 19:2023.03.15.532726. doi: 10.1101/2023.03.15.532726. bioRxiv. 2023. Update in: Elife. 2023 Dec 18;12:RP87445. doi: 10.7554/eLife.87445. PMID: 36993168 Free PMC article. Updated. Preprint.
References
-
- Adam RC, Yang H, Ge Y, Infarinato NR, Gur-Cohen S, Miao Y, Wang P, Zhao Y, Lu CP, Kim JE, Ko JY, Paik SS, Gronostajski RM, Kim J, Krueger JG, Zheng D, Fuchs E. NFI transcription factors provide chromatin access to maintain stem cell identity while preventing unintended lineage fate choices. Nature Cell Biology. 2020;22:640–650. doi: 10.1038/s41556-020-0513-0. - DOI - PMC - PubMed
-
- Bashkirova E. Bashkirova_Zonal_Or_2023. swh:1:rev:cdc45021f629ba3db70da394883c649437deea72Software Heritage. 2023 https://archive.softwareheritage.org/swh:1:dir:25923387c14d9a3f06f994abf...
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

