Short-Chain Fatty Acid Production by Gut Microbiota Predicts Treatment Response in Multiple Myeloma
- PMID: 38109212
- PMCID: PMC10870002
- DOI: 10.1158/1078-0432.CCR-23-0195
Short-Chain Fatty Acid Production by Gut Microbiota Predicts Treatment Response in Multiple Myeloma
Abstract
Purpose: The gut microbiota plays important roles in health and disease. We questioned whether the gut microbiota and related metabolites are altered in monoclonal gammopathies and evaluated their potential role in multiple myeloma and its response to treatment.
Experimental design: We used 16S rRNA sequencing to characterize and compare the gut microbiota of patients with monoclonal gammopathy of undetermined significance (n = 11), smoldering multiple myeloma (n = 9), newly diagnosed multiple myeloma (n = 11), relapsed/refractory multiple myeloma (n = 6), or with complete remission (n = 9). Short-chain fatty acids (SCFA) were quantified in serum and tested in cell lines. Relevant metabolites were validated in a second cohort of 62 patients.
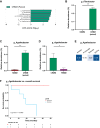
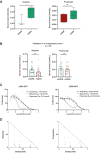
Results: Significant differences in alpha- and beta diversity were present across the groups and both were lower in patients with relapse/refractory disease and higher in patients with complete remission after treatment. Differences were found in the abundance of several microbiota taxa across disease progression and in response to treatment. Bacteria involved in SCFA production, including Prevotella, Blautia, Weissella, and Agathobacter, were more represented in the premalignant or complete remission samples, and patients with higher levels of Agathobacter showed better overall survival. Serum levels of butyrate and propionate decreased across disease progression and butyrate was positively associated with a better response. Both metabolites had antiproliferative effects in multiple myeloma cell lines.
Conclusions: We demonstrate that SCFAs metabolites and the gut microbiota associated with their production might have beneficial effects in disease evolution and response to treatment, underscoring its therapeutic potential and value as a predictor.
©2023 The Authors; Published by the American Association for Cancer Research.
Figures




References
-
- Kumar S, Paiva B, Anderson KC, Durie B, Landgren O, Moreau P, et al. . International myeloma working group consensus criteria for response and minimal residual disease assessment in multiple myeloma. Lancet Oncol 2016;17:e328–46. - PubMed
-
- van de Donk NWCJ, Pawlyn C, Yong KL. Multiple myeloma. Lancet 2021;397:410–27. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

