Hyperactive Rac stimulates cannibalism of living target cells and enhances CAR-M-mediated cancer cell killing
- PMID: 38109551
- PMCID: PMC10756302
- DOI: 10.1073/pnas.2310221120
Hyperactive Rac stimulates cannibalism of living target cells and enhances CAR-M-mediated cancer cell killing
Abstract
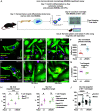
The 21kD GTPase Rac is an evolutionarily ancient regulator of cell shape and behavior. Rac2 is predominantly expressed in hematopoietic cells where it is essential for survival and motility. The hyperactivating mutation Rac2E62K also causes human immunodeficiency, although the mechanism remains unexplained. Here, we report that in Drosophila, hyperactivating Rac stimulates ovarian cells to cannibalize neighboring cells, destroying the tissue. We then show that hyperactive Rac2E62K stimulates human HL60-derived macrophage-like cells to engulf and kill living T cell leukemia cells. Primary mouse Rac2+/E62K bone-marrow-derived macrophages also cannibalize primary Rac2+/E62K T cells due to a combination of macrophage hyperactivity and T cell hypersensitivity to engulfment. Additionally, Rac2+/E62K macrophages non-autonomously stimulate wild-type macrophages to engulf T cells. Rac2E62K also enhances engulfment of target cancer cells by chimeric antigen receptor-expressing macrophages (CAR-M) in a CAR-dependent manner. We propose that Rac-mediated cell cannibalism may contribute to Rac2+/E62K human immunodeficiency and enhance CAR-M cancer immunotherapy.
Keywords: Drosophila; Rac GTPase; lymphopenia; macrophage; phagocytosis.
Conflict of interest statement
Competing interests statement:The University of California, Santa Barbara has filed a patent application on the basis of these findings.
Figures






References
-
- Ridley A. J., Paterson H. F., Johnston C. L., Diekmann D., Hall A., The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell 70, 401–410 (1992). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

