Impaired neuron differentiation in GBA-associated Parkinson's disease is linked to cell cycle defects in organoids
- PMID: 38110400
- PMCID: PMC10728202
- DOI: 10.1038/s41531-023-00616-8
Impaired neuron differentiation in GBA-associated Parkinson's disease is linked to cell cycle defects in organoids
Abstract
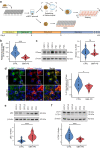
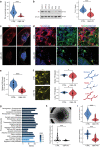
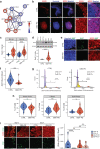
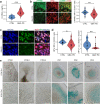
The mechanisms underlying Parkinson's disease (PD) etiology are only partially understood despite intensive research conducted in the field. Recent evidence suggests that early neurodevelopmental defects might play a role in cellular susceptibility to neurodegeneration. To study the early developmental contribution of GBA mutations in PD we used patient-derived iPSCs carrying a heterozygous N370S mutation in the GBA gene. Patient-specific midbrain organoids displayed GBA-PD relevant phenotypes such as reduction of GCase activity, autophagy impairment, and mitochondrial dysfunction. Genome-scale metabolic (GEM) modeling predicted changes in lipid metabolism which were validated with lipidomics analysis, showing significant differences in the lipidome of GBA-PD. In addition, patient-specific midbrain organoids exhibited a decrease in the number and complexity of dopaminergic neurons. This was accompanied by an increase in the neural progenitor population showing signs of oxidative stress-induced damage and premature cellular senescence. These results provide insights into how GBA mutations may lead to neurodevelopmental defects thereby predisposing to PD pathology.
© 2023. The Author(s).
Conflict of interest statement
The authors J.J., S.B. and J.C.S. declare no competing non-financial interests but declare competing financial interests as cofounders and shareholders of OrganoTherapeutics société à responsabilité limitée (SARL). The remaining authors declare no competing interests.
Figures


References
Grants and funding
- INTER/JPND/15/11092422/Fonds National de la Recherche Luxembourg (National Research Fund)
- FNR/NCER13/BM/11264123/Fonds National de la Recherche Luxembourg (National Research Fund)
- AFR/Fonds National de la Recherche Luxembourg (National Research Fund)
- FNR/NCER13/BM/11264123/Fonds National de la Recherche Luxembourg (National Research Fund)
LinkOut - more resources
Full Text Sources
Research Materials

