Mutant GGGGCC RNA prevents YY1 from binding to Fuzzy promoter which stimulates Wnt/β-catenin pathway in C9ALS/FTD
- PMID: 38110419
- PMCID: PMC10728118
- DOI: 10.1038/s41467-023-44215-w
Mutant GGGGCC RNA prevents YY1 from binding to Fuzzy promoter which stimulates Wnt/β-catenin pathway in C9ALS/FTD
Abstract
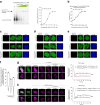
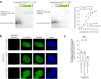
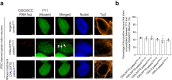
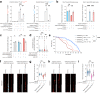
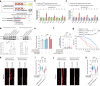
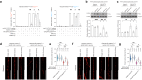
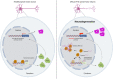
The GGGGCC hexanucleotide repeat expansion mutation in the chromosome 9 open reading frame 72 (C9orf72) gene is a major genetic cause of amyotrophic lateral sclerosis and frontotemporal dementia (C9ALS/FTD). In this study, we demonstrate that the zinc finger (ZF) transcriptional regulator Yin Yang 1 (YY1) binds to the promoter region of the planar cell polarity gene Fuzzy to regulate its transcription. We show that YY1 interacts with GGGGCC repeat RNA via its ZF and that this interaction compromises the binding of YY1 to the FuzzyYY1 promoter sites, resulting in the downregulation of Fuzzy transcription. The decrease in Fuzzy protein expression in turn activates the canonical Wnt/β-catenin pathway and induces synaptic deficits in C9ALS/FTD neurons. Our findings demonstrate a C9orf72 GGGGCC RNA-initiated perturbation of YY1-Fuzzy transcriptional control that implicates aberrant Wnt/β-catenin signalling in C9ALS/FTD-associated neurodegeneration. This pathogenic cascade provides a potential new target for disease-modifying therapy.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Research Materials

