Proteolysis-targeting chimeras with reduced off-targets
- PMID: 38110475
- PMCID: PMC10913580
- DOI: 10.1038/s41557-023-01379-8
Proteolysis-targeting chimeras with reduced off-targets
Abstract
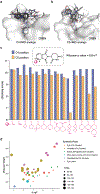
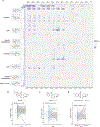
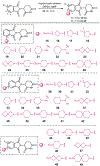
Proteolysis-targeting chimeras (PROTACs) are molecules that induce proximity between target proteins and E3 ligases triggering target protein degradation. Pomalidomide, a widely used E3 ligase recruiter in PROTACs, can independently degrade other proteins, including zinc-finger (ZF) proteins, with vital roles in health and disease. This off-target degradation hampers the therapeutic applicability of pomalidomide-based PROTACs, requiring development of PROTAC design rules that minimize off-target degradation. Here we developed a high-throughput platform that interrogates off-target degradation and found that reported pomalidomide-based PROTACs induce degradation of several ZF proteins. We generated a library of pomalidomide analogues to understand how functionalizing different positions of the phthalimide ring, hydrogen bonding, and steric and hydrophobic effects impact ZF protein degradation. Modifications of appropriate size on the C5 position reduced off-target ZF degradation, which we validated through target engagement and proteomics studies. By applying these design principles, we developed anaplastic lymphoma kinase oncoprotein-targeting PROTACs with enhanced potency and minimal off-target degradation.
© 2023. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Broad Institute has filed a patent application including the work described herein. A.C. is a founder and Scientific Advisory Board (SAB) member in Photys therapeutics. E.S.F. is a founder, SAB member, and equity holder in Civetta, Lighthorse, Proximity and Neomorph (board of directors), SAB member and equity holder in Photys and Avilar, and a consultant to Astellas, Novartis, Sanofi, Deerfield and EcoR1. The Fischer lab receives or has received research funding from Novartis, Astellas, Interline and Deerfield. D.R.L. is a consultant and co-founder of Exo Therapeutics, a company that develops small-molecule therapeutics. K.A.D. is a consultant to Kronos Bio and Neomorph Inc. The remaining authors declare no competing financial interests.
Figures







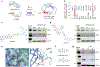




References
-
- Chamberlain PP & Cathers BE Cereblon modulators: low molecular weight inducers of protein degradation. Drug Discov. Today Technol 31, 29–34 (2019). - PubMed
-
- Kozicka Z & Thomä NH Haven’t got a glue: protein surface variation for the design of molecular glue degraders. Cell Chem. Biol 28, 1032–1047 (2021). - PubMed
-
- Ito T et al. Identification of a primary target of thalidomide teratogenicity. Science 327, 1345–1350 (2010). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

