S9.6-based hybrid capture immunoassay for pathogen detection
- PMID: 38110611
- PMCID: PMC10728093
- DOI: 10.1038/s41598-023-49881-w
S9.6-based hybrid capture immunoassay for pathogen detection
Abstract
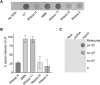
The detection of pathogens is critical for clinical diagnosis and public health surveillance. Detection is usually done with nucleic acid-based tests (NATs) and rapid antigen tests (e.g., lateral flow assays [LFAs]). Although NATs are more sensitive and specific, their use is often limited in resource-poor settings due to specialized requirements. To address this limitation, we developed a rapid DNA-RNA Hybrid Capture immunoassay (HC) that specifically detects RNA from pathogens. This assay utilizes a unique monoclonal antibody, S9.6, which binds DNA-RNA hybrids. Biotinylated single-stranded DNA probes are hybridized to target RNAs, followed by hybrid capture on streptavidin and detection with S9.6. The HC-ELISA assay can detect as few as 104 RNA molecules that are 2.2 kb in length. We also adapted this assay into a LFA format, where captured Bacillus anthracis rpoB RNA of 3.5 kb length was detectable from a bacterial load equivalent to 107 CFU per 100 mg of mouse tissue using either HC-ELISA or HC-LFA. Importantly, we also demonstrated the versatility of HC by detecting other pathogens, including SARS-CoV-2 and Toxoplasma gondii, showing its potential for broad pathogen detection. Notably, HC does not require amplification of the target nucleic acid and utilizes economical formats like ELISA and LFA, making it suitable for use in sentinel labs for pathogen detection or as a molecular tool in basic research laboratories. Our study highlights the potential of HC as a sensitive and versatile method for RNA-based pathogen detection.
© 2023. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

