The presence of a G-quadruplex prone sequence upstream of a minimal promoter increases transcriptional activity in the yeast Saccharomyces cerevisiae
- PMID: 38112096
- PMCID: PMC10730334
- DOI: 10.1042/BSR20231348
The presence of a G-quadruplex prone sequence upstream of a minimal promoter increases transcriptional activity in the yeast Saccharomyces cerevisiae
Abstract
Non-canonical secondary structures in DNA are increasingly being revealed as critical players in DNA metabolism, including modulating the accessibility and activity of promoters. These structures comprise the so-called G-quadruplexes (G4s) that are formed from sequences rich in guanine bases. Using a well-defined transcriptional reporter system, we sought to systematically investigate the impact of the presence of G4 structures on transcription in yeast Saccharomyces cerevisiae. To this aim, different G4 prone sequences were modeled to vary the chance of intramolecular G4 formation, analyzed in vitro by Thioflavin T binding test and circular dichroism and then placed at the yeast ADE2 locus on chromosome XV, downstream and adjacent to a P53 response element (RE) and upstream from a minimal CYC1 promoter and Luciferase 1 (LUC1) reporter gene in isogenic strains. While the minimal CYC1 promoter provides basal reporter activity, the P53 RE enables LUC1 transactivation under the control of P53 family proteins expressed under the inducible GAL1 promoter. Thus, the impact of the different G4 prone sequences on both basal and P53 family protein-dependent expression was measured after shifting cells onto galactose containing medium. The results showed that the presence of G4 prone sequences upstream of a yeast minimal promoter increased its basal activity proportionally to their potential to form intramolecular G4 structures; consequently, this feature, when present near the target binding site of P53 family transcription factors, can be exploited to regulate the transcriptional activity of P53, P63 and P73 proteins.
Keywords: G-quadruplex; p53; transcriptional activity; yeast.
© 2023 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures

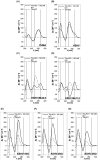
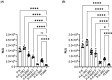


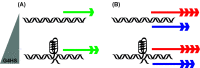
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

