FlhF affects the subcellular clustering of WspR through HsbR in Pseudomonas aeruginosa
- PMID: 38112425
- PMCID: PMC10807432
- DOI: 10.1128/aem.01548-23
FlhF affects the subcellular clustering of WspR through HsbR in Pseudomonas aeruginosa
Abstract
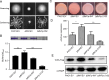
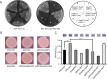
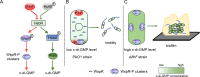
In bacteria, the second messenger cyclic di-GMP (c-di-GMP) is synthesized and degraded by multiple diguanylate cyclases (DGCs) and phosphodiesterases. A high level of c-di-GMP induces biofilm formation and represses motility. WspR, a hybrid response regulator DGC, produces c-di-GMP when it is phosphorylated. FlhF, a signal recognition particle-type GTPase, is initially localized to the cell poles and is indispensable for polar flagellar localization in Pseudomonas aeruginosa. In this study, we report that deletion of flhF affected biofilm formation and the c-di-GMP level in P. aeruginosa. Phenotypic analysis of a flhF knockout mutant revealed increased biofilm formation, wrinkled colonies on Congo red agar, and an elevated c-di-GMP level compared to the wild-type strain, PAO1. Yeast and bacterial two-hybrid systems showed that FlhF binds to the response regulator HsbR, and HsbR binds to WspR. Deletion of hsbR or wspR in the ΔflhF background abolished the phenotype of ΔflhF. In addition, confocal microscopy demonstrated that WspR-GFP was distributed throughout the cytoplasm and formed a visible cluster at one cell pole in PAO1 and ΔhsbR, but it was mainly distributed as visible clusters at the lateral side of the periplasm and with visible clusters at both cell poles in ΔflhF. These findings suggest that FlhF influences the subcellular cluster and localization of WspR and negatively modulates WspR DGC activity in a manner dependent on HsbR. Together, our findings demonstrate a novel mechanism for FlhF modulating the lifestyle transition between motility and biofilm via HsbR to regulate the DGC activity of WspR.IMPORTANCECyclic di-GMP (c-di-GMP) is a second messenger that controls flagellum biosynthesis, adhesion, virulence, motility, exopolysaccharide production, and biofilm formation in bacteria. Recent research has shown that distinct diguanylate cyclases (DGCs) or phosphodiesterases (PDEs) produce highly specific outputs. Some DGCs and PDEs contribute to the total global c-di-GMP concentration, but others only affect local c-di-GMP in a microenvironment. However, the underlying mechanisms are unclear. Here, we report that FlhF affects the localization and DGC activity of WspR via HsbR and is implicated in local c-di-GMP signaling in Pseudomonas aeruginosa. This study establishes the link between the c-di-GMP signaling system and the flagellar localization and provides insight for understanding the complex regulatory network of c-di-GMP signaling.
Keywords: FlhF; WspR; biofilm formation; c-di-GMP; subcellular clustering.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
The Pseudomonas aeruginosa diguanylate cyclase GcbA, a homolog of P. fluorescens GcbA, promotes initial attachment to surfaces, but not biofilm formation, via regulation of motility.J Bacteriol. 2014 Aug;196(15):2827-41. doi: 10.1128/JB.01628-14. Epub 2014 Jun 2. J Bacteriol. 2014. PMID: 24891445 Free PMC article.
-
Subcellular clustering of the phosphorylated WspR response regulator protein stimulates its diguanylate cyclase activity.mBio. 2013 May 7;4(3):e00242-13. doi: 10.1128/mBio.00242-13. mBio. 2013. PMID: 23653447 Free PMC article.
-
The c-di-GMP Phosphodiesterase PipA (PA0285) Regulates Autoaggregation and Pf4 Bacteriophage Production in Pseudomonas aeruginosa PAO1.Appl Environ Microbiol. 2022 Jun 28;88(12):e0003922. doi: 10.1128/aem.00039-22. Epub 2022 May 31. Appl Environ Microbiol. 2022. PMID: 35638845 Free PMC article.
-
c-di-GMP signaling in Pseudomonas syringae complex.Microbiol Res. 2023 Oct;275:127445. doi: 10.1016/j.micres.2023.127445. Epub 2023 Jul 6. Microbiol Res. 2023. PMID: 37450986 Review.
-
Investigating c-di-GMP Signaling in Vibrio parahaemolyticus: Biological Effects and Mechanisms of Regulation.Curr Microbiol. 2025 Jun 2;82(7):319. doi: 10.1007/s00284-025-04260-8. Curr Microbiol. 2025. PMID: 40455311 Review.
Cited by
-
Genetic analysis of flagellar-mediated surface sensing by Pseudomonas aeruginosa PA14.J Bacteriol. 2025 Jul 24;207(7):e0052024. doi: 10.1128/jb.00520-24. Epub 2025 Jun 5. J Bacteriol. 2025. PMID: 40470954 Free PMC article.
-
Echinacoside reduces intracellular c-di-GMP levels and potentiates tobramycin activity against Pseudomonas aeruginosa biofilm aggregates.NPJ Biofilms Microbiomes. 2025 Mar 7;11(1):40. doi: 10.1038/s41522-025-00673-2. NPJ Biofilms Microbiomes. 2025. PMID: 40055321 Free PMC article.
-
Genetic Analysis of Flagellar-Mediated Surface Sensing by Pseudomonas aeruginosa PA14.bioRxiv [Preprint]. 2024 Dec 5:2024.12.05.627040. doi: 10.1101/2024.12.05.627040. bioRxiv. 2024. Update in: J Bacteriol. 2025 Jul 24;207(7):e0052024. doi: 10.1128/jb.00520-24. PMID: 39677620 Free PMC article. Updated. Preprint.
References
-
- Høiby N, Bjarnsholt T, Moser C, Bassi GL, Coenye T, Donelli G, Hall-Stoodley L, Holá V, Imbert C, Kirketerp-Møller K, Lebeaux D, Oliver A, Ullmann AJ, Williams C, ESCMID Study Group for Biofilms and Consulting External Expert Werner Zimmerli . 2015. ESCMID guideline for the diagnosis and treatment of biofilm infections 2014. Clin Microbiol Infect 21 Suppl 1:S1–25. doi:10.1016/j.cmi.2014.10.024 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

