Requirement of GrgA for Chlamydia infectious progeny production, optimal growth, and efficient plasmid maintenance
- PMID: 38112466
- PMCID: PMC10790707
- DOI: 10.1128/mbio.02036-23
Requirement of GrgA for Chlamydia infectious progeny production, optimal growth, and efficient plasmid maintenance
Abstract
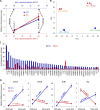
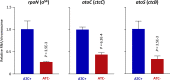
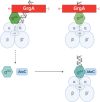
Hallmarks of the developmental cycle of the obligate intracellular pathogenic bacterium Chlamydia are the primary differentiation of the infectious elementary body (EB) into the proliferative reticulate body (RB) and the secondary differentiation of RBs back into EBs. The mechanisms regulating these transitions remain unclear. In this report, we developed an effective novel strategy termed dependence on plasmid-mediated expression (DOPE) that allows for the knockdown of essential genes in Chlamydia. We demonstrate that GrgA, a Chlamydia-specific transcription factor, is essential for the secondary differentiation and optimal growth of RBs. We also show that GrgA, a chromosome-encoded regulatory protein, controls the maintenance of the chlamydial virulence plasmid. Transcriptomic analysis further indicates that GrgA functions as a critical regulator of all three sigma factors that recognize different promoter sets at developmental stages. The DOPE strategy outlined here should provide a valuable tool for future studies examining chlamydial growth, development, and pathogenicity.
Keywords: Chlamydia; GrgA; regulation of gene expression; transcription factors; transcriptional regulation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Update of
-
Requirement of GrgA for Chlamydia infectious progeny production, optimal growth, and efficient plasmid maintenance.bioRxiv [Preprint]. 2023 Aug 2:2023.08.02.551707. doi: 10.1101/2023.08.02.551707. bioRxiv. 2023. Update in: mBio. 2024 Jan 16;15(1):e0203623. doi: 10.1128/mbio.02036-23. PMID: 37577610 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
