The chromosome-scale genome and the genetic resistance machinery against insect herbivores of the Mexican toloache, Datura stramonium
- PMID: 38113048
- PMCID: PMC10849327
- DOI: 10.1093/g3journal/jkad288
The chromosome-scale genome and the genetic resistance machinery against insect herbivores of the Mexican toloache, Datura stramonium
Abstract
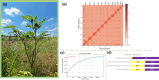
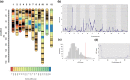
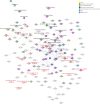
Plant resistance refers to the heritable ability of plants to reduce damage caused by natural enemies, such as herbivores and pathogens, either through constitutive or induced traits like chemical compounds or trichomes. However, the genetic architecture-the number and genome location of genes that affect plant defense and the magnitude of their effects-of plant resistance to arthropod herbivores in natural populations remains poorly understood. In this study, we aimed to unveil the genetic architecture of plant resistance to insect herbivores in the annual herb Datura stramonium (Solanaceae) through quantitative trait loci mapping. We achieved this by assembling the species' genome and constructing a linkage map using an F2 progeny transplanted into natural habitats. Furthermore, we conducted differential gene expression analysis between undamaged and damaged plants caused by the primary folivore, Lema daturaphila larvae. Our genome assembly resulted in 6,109 scaffolds distributed across 12 haploid chromosomes. A single quantitative trait loci region on chromosome 3 was associated with plant resistance, spanning 0 to 5.17 cM. The explained variance by the quantitative trait loci was 8.44%. Our findings imply that the resistance mechanisms of D. stramonium are shaped by the complex interplay of multiple genes with minor effects. Protein-protein interaction networks involving genes within the quantitative trait loci region and overexpressed genes uncovered the key role of receptor-like cytoplasmic kinases in signaling and regulating tropane alkaloids and terpenoids, which serve as powerful chemical defenses against D. stramonium herbivores. The data generated in our study constitute important resources for delving into the evolution and ecology of secondary compounds mediating plant-insect interactions.
Keywords: Plant Genetics and Genomics; Solanaceae; genomics of plant defense; herbivory; plant resistance; quantitative trait loci (QTLs) of plant resistance.
© The Author(s) 2023. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest The authors declare no conflicts of interest.
Figures


Similar articles
-
Genomic signatures of the evolution of defence against its natural enemies in the poisonous and medicinal plant Datura stramonium (Solanaceae).Sci Rep. 2021 Jan 13;11(1):882. doi: 10.1038/s41598-020-79194-1. Sci Rep. 2021. PMID: 33441607 Free PMC article.
-
Genomic and chemical evidence for local adaptation in resistance to different herbivores in Datura stramonium.Evolution. 2020 Dec;74(12):2629-2643. doi: 10.1111/evo.14097. Epub 2020 Oct 5. Evolution. 2020. PMID: 32935854
-
A Trip Back Home: Resistance to Herbivores of Native and Non-Native Plant Populations of Datura stramonium.Plants (Basel). 2024 Jan 2;13(1):131. doi: 10.3390/plants13010131. Plants (Basel). 2024. PMID: 38202439 Free PMC article.
-
Genetic Warfare: The Plant Genome's Role in Fending Off Insect Invaders.Arch Insect Biochem Physiol. 2024 Dec;117(4):e70021. doi: 10.1002/arch.70021. Arch Insect Biochem Physiol. 2024. PMID: 39726337 Review.
-
Mechanisms of plant defense against insect herbivores.Plant Signal Behav. 2012 Oct 1;7(10):1306-20. doi: 10.4161/psb.21663. Epub 2012 Aug 20. Plant Signal Behav. 2012. PMID: 22895106 Free PMC article. Review.
References
-
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data [Online]. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
-
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 57(1):289–300. doi:10.1111/j.2517-6161.1995.tb02031.x. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
