Tiam1 methylation by NSD2 promotes Rac1 signaling activation and colon cancer metastasis
- PMID: 38113258
- PMCID: PMC10756287
- DOI: 10.1073/pnas.2305684120
Tiam1 methylation by NSD2 promotes Rac1 signaling activation and colon cancer metastasis
Abstract
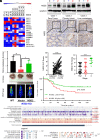
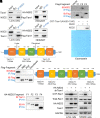
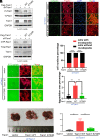
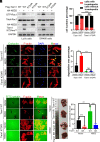
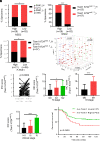
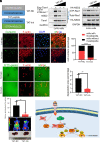
Metastasis is a major cause of cancer therapy failure and mortality. However, targeting metastatic seeding and colonization remains a significant challenge. In this study, we identified NSD2, a histone methyltransferase responsible for dimethylating histone 3 at lysine 36, as being overexpressed in metastatic tumors. Our findings suggest that NSD2 overexpression enhances tumor metastasis both in vitro and in vivo. Further analysis revealed that NSD2 promotes tumor metastasis by activating Rac1 signaling. Mechanistically, NSD2 combines with and activates Tiam1 (T lymphoma invasion and metastasis 1) and promotes Rac1 signaling by methylating Tiam1 at K724. In vivo and in vitro studies revealed that Tiam1 K724 methylation could be a predictive factor for cancer prognosis and a potential target for metastasis inhibition. Furthermore, we have developed inhibitory peptide which was proved to inhibit tumor metastasis through blocking the interaction between NSD2 and Tiam1. Our results demonstrate that NSD2-methylated Tiam1 promotes Rac1 signaling and cancer metastasis. These results provide insights into the inhibition of tumor metastasis.
Keywords: NSD2; Rac1 signaling; Tiam1 methylation; cancer metastasis.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures

References
-
- Bray F., et al. , Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 (2018). - PubMed
-
- Chen W., et al. , Cancer statistics in China, 2015. CA Cancer J. Clin. 66, 115–132 (2016). - PubMed
-
- Siegel R. L., Miller K. D., Jemal A., Cancer statistics, 2020. CA Cancer J. Clin. 70, 7–30 (2020). - PubMed
-
- Douillard J. Y., et al. , Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N. Engl. J. Med. 369, 1023–1034 (2013). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

