pyCapsid: identifying dominant dynamics and quasi-rigid mechanical units in protein shells
- PMID: 38113434
- PMCID: PMC10786678
- DOI: 10.1093/bioinformatics/btad761
pyCapsid: identifying dominant dynamics and quasi-rigid mechanical units in protein shells
Abstract
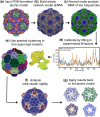
Summary: pyCapsid is a Python package developed to facilitate the characterization of the dynamics and quasi-rigid mechanical units of protein shells and other protein complexes. The package was developed in response to the rapid increase of high-resolution structures, particularly capsids of viruses, requiring multiscale biophysical analyses. Given a protein shell, pyCapsid generates the collective vibrations of its amino-acid residues, identifies quasi-rigid mechanical regions associated with the disassembly of the structure, and maps the results back to the input proteins for interpretation. pyCapsid summarizes the main results in a report that includes publication-quality figures.
Availability and implementation: pyCapsid's source code is available under MIT License on GitHub. It is compatible with Python 3.8-3.10 and has been deployed in two leading Python package-management systems, PIP and Conda. Installation instructions and tutorials are available in the online documentation and in the pyCapsid's YouTube playlist. In addition, a cloud-based implementation of pyCapsid is available as a Google Colab notebook. pyCapsid Colab does not require installation and generates the same report and outputs as the installable version. Users can post issues regarding pyCapsid in the repository's issues section.
© The Author(s) 2023. Published by Oxford University Press.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
CoMut: visualizing integrated molecular information with comutation plots.Bioinformatics. 2020 Aug 1;36(15):4348-4349. doi: 10.1093/bioinformatics/btaa554. Bioinformatics. 2020. PMID: 32502231 Free PMC article.
-
PHi-C2: interpreting Hi-C data as the dynamic 3D genome state.Bioinformatics. 2022 Oct 31;38(21):4984-4986. doi: 10.1093/bioinformatics/btac613. Bioinformatics. 2022. PMID: 36087002 Free PMC article.
-
SBMLDiagrams: a python package to process and visualize SBML layout and render.Bioinformatics. 2023 Jan 1;39(1):btac730. doi: 10.1093/bioinformatics/btac730. Bioinformatics. 2023. PMID: 36370074 Free PMC article.
-
HOMELETTE: a unified interface to homology modelling software.Bioinformatics. 2022 Mar 4;38(6):1749-1751. doi: 10.1093/bioinformatics/btab866. Bioinformatics. 2022. PMID: 34954790 Free PMC article.
-
danRerLib: a Python package for zebrafish transcriptomics.Bioinform Adv. 2024 May 6;4(1):vbae065. doi: 10.1093/bioadv/vbae065. eCollection 2024. Bioinform Adv. 2024. PMID: 38770229 Free PMC article.
Cited by
-
Theoretical Studies on Assembly, Physical Stability, and Dynamics of Viruses.Subcell Biochem. 2024;105:693-741. doi: 10.1007/978-3-031-65187-8_19. Subcell Biochem. 2024. PMID: 39738961 Review.
-
Stabilization mechanism accommodating genome length variation in evolutionarily related viral capsids.Nat Commun. 2025 Apr 2;16(1):3145. doi: 10.1038/s41467-025-58298-0. Nat Commun. 2025. PMID: 40175362 Free PMC article.
References
-
- Agirre J, Goret G, LeGoff M. et al. Cryo-electron microscopy reconstructions of triatoma virus particles: a clue to unravel genome delivery and capsid disassembly. J Gen Virol 2013;94:1058–68. - PubMed

