ELISL: early-late integrated synthetic lethality prediction in cancer
- PMID: 38113447
- PMCID: PMC11616771
- DOI: 10.1093/bioinformatics/btad764
ELISL: early-late integrated synthetic lethality prediction in cancer
Abstract
Motivation: Anti-cancer therapies based on synthetic lethality (SL) exploit tumour vulnerabilities for treatment with reduced side effects, by targeting a gene that is jointly essential with another whose function is lost. Computational prediction is key to expedite SL screening, yet existing methods are vulnerable to prevalent selection bias in SL data and reliant on cancer or tissue type-specific omics, which can be scarce. Notably, sequence similarity remains underexplored as a proxy for related gene function and joint essentiality.
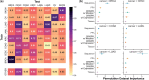
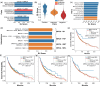
Results: We propose ELISL, Early-Late Integrated SL prediction with forest ensembles, using context-free protein sequence embeddings and context-specific omics from cell lines and tissue. Across eight cancer types, ELISL showed superior robustness to selection bias and recovery of known SL genes, as well as promising cross-cancer predictions. Co-occurring mutations in a BRCA gene and ELISL-predicted pairs from the HH, FGF, WNT, or NEIL gene families were associated with longer patient survival times, revealing therapeutic potential.
Availability and implementation: Data: 10.6084/m9.figshare.23607558 & Code: github.com/joanagoncalveslab/ELISL.
© The Author(s) 2023. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures


References
-
- Ashworth A, Lord CJ.. Synthetic lethal therapies for cancer: what’s next after PARP inhibitors? Nat Rev Clin Oncol 2018;15:564–76. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

