Circulating biomarkers in familial cerebral cavernous malformation
- PMID: 38113759
- PMCID: PMC10767159
- DOI: 10.1016/j.ebiom.2023.104914
Circulating biomarkers in familial cerebral cavernous malformation
Abstract
Background: Cerebral Cavernous Malformation (CCM) is a rare cerebrovascular disease, characterized by the presence of multiple vascular malformations that may result in intracerebral hemorrhages (ICHs), seizure(s), or focal neurological deficits (FND). Familial CCM (fCCM) is due to loss of function mutations in one of the three independent genes KRIT1 (CCM1), Malcavernin (CCM2), or Programmed Cell death 10 (PDCD10/CCM3). The aim of this study was to identify plasma protein biomarkers of fCCM to assess the severity of the disease and predict its progression.
Methods: Here, we have investigated plasma samples derived from n = 71 symptomatic fCCM patients (40 female/31 male) and n = 17 healthy donors (HD) (9 female/8 male) of the Phase 1/2 Treat_CCM trial, using multiplexed protein profiling approaches.
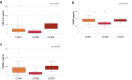
Findings: Biomarkers as sCD14 (p = 0.00409), LBP (p = 0.02911), CXCL4 (p = 0.038), ICAM-1 (p = 0.02013), ANG2 (p = 0.026), CCL5 (p = 0.00403), THBS1 (p = 0.0043), CRP (p = 0.0092), and HDL (p = 0.027), were significantly different in fCCM compared to HDs. Of note, sENG (p = 0.011), THBS1 (p = 0.011) and CXCL4 (p = 0.011), were correlated to CCM genotype. sROBO4 (p = 0.014), TM (p = 0.026) and CRP (p = 0.040) were able to predict incident adverse clinical events, such as ICH, FND or seizure. GDF-15, FLT3L, CXCL9, FGF-21 and CDCP1, were identified as predictors of the formation of new MRI-detectable lesions over 2-year follow-up. Furthermore, the functional relevance of ang2, thbs1, robo4 and cdcp1 markers was validated by zebrafish pre-clinical model of fCCM.
Interpretation: Overall, our study identifies a set of biochemical parameters to predict CCM progression, suggesting biological interpretations and potential therapeutic approaches to CCM disease.
Funding: Italian Medicines Agency, Associazione Italiana per la Ricerca sul Cancro (AIRC), ERC, Leducq Transatlantic Network of Excellence, Swedish Research Council.
Keywords: Biomarkers; Familial cerebral cavernous malformation; Proteomics; Vascular biology.
Copyright © 2023 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests B.B is inventor of patents on PTX3 and obtains royalties on PTX3-related reagents. N.B. serves on Advisory Board and Speakers Bureau for Celgene and Janssen, and on Speakers Bureau for Takeda, Amgen.
Figures



References
-
- Cox E.M., Bambakidis N.C., Cohen M.L. Pathology of cavernous malformations. Handb Clin Neurol. 2017;143:267–277. - PubMed
-
- Kivelev J., Niemelä M., Hernesniemi J. Treatment strategies in cavernomas of the brain and spine. J Clin Neurosci. 2012;19:491–497. - PubMed
-
- Fischer A., Zalvide J., Faurobert E., Albiges-Rizo C., Tournier-Lasserve E. Cerebral cavernous malformations: from CCM genes to endothelial cell homeostasis. Trends Mol Med. 2013;19:302–308. - PubMed
-
- Labauge P., Denier C., Bergametti F., Tournier-Lasserve E. Genetics of cavernous angiomas. Lancet Neurol. 2007;6:237–244. - PubMed
-
- Lampugnani M.G., Malinverno M., Dejana E., Rudini N. Endothelial cell disease: emerging knowledge from cerebral cavernous malformations. Curr Opin Hematol. 2017;24:256–264. - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

