Identification of miRNA858 long-loop precursors in seed plants
- PMID: 38114096
- PMCID: PMC11062470
- DOI: 10.1093/plcell/koad315
Identification of miRNA858 long-loop precursors in seed plants
Abstract
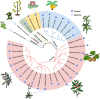
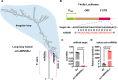
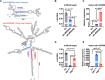
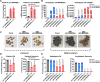
MicroRNAs (miRNAs) are a class of nonprotein-coding short transcripts that provide a layer of post-transcriptional regulation essential to many plant biological processes. MiR858, which targets the transcripts of MYB transcription factors, can affect a range of secondary metabolic processes. Although miR858 and its 187-nt precursor have been well studied in Arabidopsis (Arabidopsis thaliana), a systematic investigation of miR858 precursors and their functions across plant species is lacking due to a problem in identifying the transcripts that generate this subclass. By re-evaluating the transcript of miR858 and relaxing the length cut-off for identifying hairpins, we found in kiwifruit (Actinidia chinensis) that miR858 has long-loop hairpins (1,100 to 2,100 nt), whose intervening sequences between miRNA generating complementary sites were longer than all previously reported miRNA hairpins. Importantly, these precursors of miR858 containing long-loop hairpins (termed MIR858L) are widespread in seed plants including Arabidopsis, varying between 350 and 5,500 nt. Moreover, we showed that MIR858L has a greater impact on proanthocyanidin and flavonol levels in both Arabidopsis and kiwifruit. We suggest that an active MIR858L-MYB regulatory module appeared in the transition of early land plants to large upright flowering plants, making a key contribution to plant secondary metabolism.
© The Author(s) 2023. Published by Oxford University Press on behalf of American Society of Plant Biologists. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Conflict of interest statement
Conflict of interest statement. The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

