Efficient generation of a self-organizing neuromuscular junction model from human pluripotent stem cells
- PMID: 38114482
- PMCID: PMC10730704
- DOI: 10.1038/s41467-023-43781-3
Efficient generation of a self-organizing neuromuscular junction model from human pluripotent stem cells
Abstract
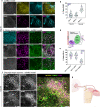
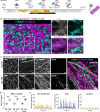
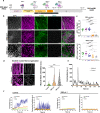
The complex neuromuscular network that controls body movements is the target of severe diseases that result in paralysis and death. Here, we report the development of a robust and efficient self-organizing neuromuscular junction (soNMJ) model from human pluripotent stem cells that can be maintained long-term in simple adherent conditions. The timely application of specific patterning signals instructs the simultaneous development and differentiation of position-specific brachial spinal neurons, skeletal muscles, and terminal Schwann cells. High-content imaging reveals self-organized bundles of aligned muscle fibers surrounded by innervating motor neurons that form functional neuromuscular junctions. Optogenetic activation and pharmacological interventions show that the spinal neurons actively instruct the synchronous skeletal muscle contraction. The generation of a soNMJ model from spinal muscular atrophy patient-specific iPSCs reveals that the number of NMJs and muscle contraction is severely affected, resembling the patient's pathology. In the future, the soNMJ model could be used for high-throughput studies in disease modeling and drug development. Thus, this model will allow us to address unmet needs in the neuromuscular disease field.
© 2023. The Author(s).
Conflict of interest statement
MG and AU are co-inventors in patent applications related to the generation of a self-organizing neuromuscular junction cell culture model filed in USA (18/335,765); Singapore (10202301703 V); Canada (P4410CA). The remaining authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

