A type VII-secreted lipase toxin with reverse domain arrangement
- PMID: 38114483
- PMCID: PMC10730906
- DOI: 10.1038/s41467-023-44221-y
A type VII-secreted lipase toxin with reverse domain arrangement
Abstract
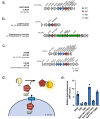
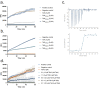
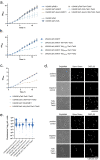
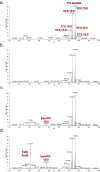
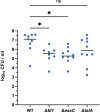
The type VII protein secretion system (T7SS) is found in many Gram-positive bacteria and in pathogenic mycobacteria. All T7SS substrate proteins described to date share a common helical domain architecture at the N-terminus that typically interacts with other helical partner proteins, forming a composite signal sequence for targeting to the T7SS. The C-terminal domains are functionally diverse and in Gram-positive bacteria such as Staphylococcus aureus often specify toxic anti-bacterial activity. Here we describe the first example of a class of T7 substrate, TslA, that has a reverse domain organisation. TslA is widely found across Bacillota including Staphylococcus, Enterococcus and Listeria. We show that the S. aureus TslA N-terminal domain is a phospholipase A with anti-staphylococcal activity that is neutralised by the immunity lipoprotein TilA. Two small helical partner proteins, TlaA1 and TlaA2 are essential for T7-dependent secretion of TslA and at least one of these interacts with the TslA C-terminal domain to form a helical stack. Cryo-EM analysis of purified TslA complexes indicate that they share structural similarity with canonical T7 substrates. Our findings suggest that the T7SS has the capacity to recognise a secretion signal present at either end of a substrate.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

