Biophysical basis of filamentous phage tactoid-mediated antibiotic tolerance in P. aeruginosa
- PMID: 38114502
- PMCID: PMC10730611
- DOI: 10.1038/s41467-023-44160-8
Biophysical basis of filamentous phage tactoid-mediated antibiotic tolerance in P. aeruginosa
Abstract
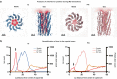
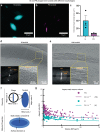
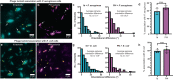
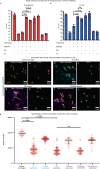
Inoviruses are filamentous phages infecting numerous prokaryotic phyla. Inoviruses can self-assemble into mesoscale structures with liquid-crystalline order, termed tactoids, which protect bacterial cells in Pseudomonas aeruginosa biofilms from antibiotics. Here, we investigate the structural, biophysical, and protective properties of tactoids formed by the P. aeruginosa phage Pf4 and Escherichia coli phage fd. A cryo-EM structure of the capsid from fd revealed distinct biochemical properties compared to Pf4. Fd and Pf4 formed tactoids with different morphologies that arise from differing phage geometries and packing densities, which in turn gave rise to different tactoid emergent properties. Finally, we showed that tactoids formed by either phage protect rod-shaped bacteria from antibiotic treatment, and that direct association with a tactoid is required for protection, demonstrating the formation of a diffusion barrier by the tactoid. This study provides insights into how filamentous molecules protect bacteria from extraneous substances in biofilms and in host-associated infections.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

