Analysis of off-tumour toxicities of T-cell-engaging bispecific antibodies via donor-matched intestinal organoids and tumouroids
- PMID: 38114742
- PMCID: PMC11087266
- DOI: 10.1038/s41551-023-01156-5
Analysis of off-tumour toxicities of T-cell-engaging bispecific antibodies via donor-matched intestinal organoids and tumouroids
Abstract
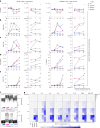
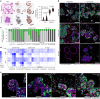
Predicting the toxicity of cancer immunotherapies preclinically is challenging because models of tumours and healthy organs do not typically fully recapitulate the expression of relevant human antigens. Here we show that patient-derived intestinal organoids and tumouroids supplemented with immune cells can be used to study the on-target off-tumour toxicities of T-cell-engaging bispecific antibodies (TCBs), and to capture clinical toxicities not predicted by conventional tissue-based models as well as inter-patient variabilities in TCB responses. We analysed the mechanisms of T-cell-mediated damage of neoplastic and donor-matched healthy epithelia at a single-cell resolution using multiplexed immunofluorescence. We found that TCBs that target the epithelial cell-adhesion molecule led to apoptosis in healthy organoids in accordance with clinical observations, and that apoptosis is associated with T-cell activation, cytokine release and intra-epithelial T-cell infiltration. Conversely, tumour organoids were more resistant to damage, probably owing to a reduced efficiency of T-cell infiltration within the epithelium. Patient-derived intestinal organoids can aid the study of immune-epithelial interactions as well as the preclinical and clinical development of cancer immunotherapies.
© 2023. The Author(s).
Conflict of interest statement
All authors, except for C.E., K.G.-J and S.P., are employees of Hoffmann-LaRoche. The company provided support in the form of salaries for authors, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. C.E., K.G.-J and S.P. declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

