Evaluation of noninvasive biospecimens for transcriptome studies
- PMID: 38114913
- PMCID: PMC10729488
- DOI: 10.1186/s12864-023-09875-4
Evaluation of noninvasive biospecimens for transcriptome studies
Abstract
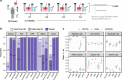
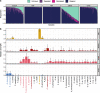
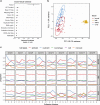
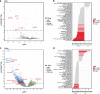
Transcriptome studies disentangle functional mechanisms of gene expression regulation and may elucidate the underlying biology of disease processes. However, the types of tissues currently collected typically assay a single post-mortem timepoint or are limited to investigating cell types found in blood. Noninvasive tissues may improve disease-relevant discovery by enabling more complex longitudinal study designs, by capturing different and potentially more applicable cell types, and by increasing sample sizes due to reduced collection costs and possible higher enrollment from vulnerable populations. Here, we develop methods for sampling noninvasive biospecimens, investigate their performance across commercial and in-house library preparations, characterize their biology, and assess the feasibility of using noninvasive tissues in a multitude of transcriptomic applications. We collected buccal swabs, hair follicles, saliva, and urine cell pellets from 19 individuals over three to four timepoints, for a total of 300 unique biological samples, which we then prepared with replicates across three library preparations, for a final tally of 472 transcriptomes. Of the four tissues we studied, we found hair follicles and urine cell pellets to be most promising due to the consistency of sample quality, the cell types and expression profiles we observed, and their performance in disease-relevant applications. This is the first study to thoroughly delineate biological and technical features of noninvasive samples and demonstrate their use in a wide array of transcriptomic and clinical analyses. We anticipate future use of these biospecimens will facilitate discovery and development of clinical applications.
Keywords: EQTLs; Hair follicles; Methods; Noninvasive RNA-sequencing; Transcriptomics; Urine cell pellets.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Collection of biospecimens from parent-child dyads in a community garden-based nutrition intervention: protocol and feasibility.BMC Nutr. 2022 Dec 5;8(1):141. doi: 10.1186/s40795-022-00640-6. BMC Nutr. 2022. PMID: 36471397 Free PMC article.
-
Single-cell transcriptome analysis of endometrial tissue.Hum Reprod. 2016 Apr;31(4):844-53. doi: 10.1093/humrep/dew008. Epub 2016 Feb 13. Hum Reprod. 2016. PMID: 26874359 Free PMC article.
-
Variation in RNA-Seq transcriptome profiles of peripheral whole blood from healthy individuals with and without globin depletion.PLoS One. 2014 Mar 7;9(3):e91041. doi: 10.1371/journal.pone.0091041. eCollection 2014. PLoS One. 2014. PMID: 24608128 Free PMC article.
-
Computational solutions for spatial transcriptomics.Comput Struct Biotechnol J. 2022 Sep 1;20:4870-4884. doi: 10.1016/j.csbj.2022.08.043. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 36147664 Free PMC article. Review.
-
Single-cell network biology for resolving cellular heterogeneity in human diseases.Exp Mol Med. 2020 Nov;52(11):1798-1808. doi: 10.1038/s12276-020-00528-0. Epub 2020 Nov 26. Exp Mol Med. 2020. PMID: 33244151 Free PMC article. Review.
Cited by
-
A map of blood regulatory variation in South Africans enables GWAS interpretation.Nat Genet. 2025 Jul;57(7):1628-1637. doi: 10.1038/s41588-025-02223-0. Epub 2025 Jun 11. Nat Genet. 2025. PMID: 40500424 Free PMC article.
-
Increasing diversity of functional genetics studies to advance biological discovery and human health.Am J Hum Genet. 2023 Dec 7;110(12):1996-2002. doi: 10.1016/j.ajhg.2023.10.012. Epub 2023 Nov 22. Am J Hum Genet. 2023. PMID: 37995684 Free PMC article.
-
Genome-wide DNA methylation and transcriptome sequencing analyses of lens tissue in an age-related mouse cataract model.PLoS One. 2025 Jan 30;20(1):e0316766. doi: 10.1371/journal.pone.0316766. eCollection 2025. PLoS One. 2025. PMID: 39883715 Free PMC article.
-
Promoter Deletion Leading to Allele Specific Expression in a Genetically Unsolved Case of Primary Ciliary Dyskinesia.Am J Med Genet A. 2025 Feb;197(2):e63880. doi: 10.1002/ajmg.a.63880. Epub 2024 Oct 4. Am J Med Genet A. 2025. PMID: 39364610
-
Saliva as a potential diagnostic medium: DNA methylation biomarkers for disorders beyond the oral cavity.NPJ Genom Med. 2025 Jun 20;10(1):49. doi: 10.1038/s41525-025-00509-0. NPJ Genom Med. 2025. PMID: 40541940 Free PMC article.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

