Single-cell and spatial transcriptomics reveal POSTN+ cancer-associated fibroblasts correlated with immune suppression and tumour progression in non-small cell lung cancer
- PMID: 38115703
- PMCID: PMC10731139
- DOI: 10.1002/ctm2.1515
Single-cell and spatial transcriptomics reveal POSTN+ cancer-associated fibroblasts correlated with immune suppression and tumour progression in non-small cell lung cancer
Abstract
Background: Cancer-associated fibroblasts (CAFs) are potential targets for cancer therapy. Due to the heterogeneity of CAFs, the influence of CAF subpopulations on the progression of lung cancer is still unclear, which impedes the translational advances in targeting CAFs.
Methods: We performed single-cell RNA sequencing (scRNA-seq) on tumour, paired tumour-adjacent, and normal samples from 16 non-small cell lung cancer (NSCLC) patients. CAF subpopulations were analyzed after integration with published NSCLC scRNA-seq data. SpaTial enhanced resolution omics-sequencing (Stereo-seq) was applied in tumour and tumour-adjacent samples from seven NSCLC patients to map the architecture of major cell populations in tumour microenvironment (TME). Immunohistochemistry (IHC) and multiplexed IHC (mIHC) were used to validate marker gene expression and the association of CAFs with immune infiltration in TME.
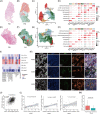
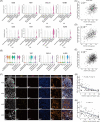
Results: A subcluster of myofibroblastic CAFs, POSTN+ CAFs, were significantly enriched in advanced tumours and presented gene expression signatures related to extracellular matrix remodeling, tumour invasion pathways and immune suppression. Stereo-seq and mIHC demonstrated that POSTN+ CAFs were in close localization with SPP1+ macrophages and were associated with the exhausted phenotype and lower infiltration of T cells. POSTN expression or the abundance of POSTN+ CAFs were associated with poor prognosis of NSCLC.
Conclusions: Our study identified a myofibroblastic CAF subpopulation, POSTN+ CAFs, which might associate with SPP1+ macrophages to promote the formation of desmoplastic architecture and participate in immune suppression. Furthermore, we showed that POSTN+ CAFs associated with cancer progression and poor clinical outcomes and may provide new insights on the treatment of NSCLC.
Keywords: POSTN; non-small cell lung cancer; single-cell RNA sequencing; spatial transcriptomics.
© 2023 The Authors. Clinical and Translational Medicine published by John Wiley & Sons Australia, Ltd on behalf of Shanghai Institute of Clinical Bioinformatics.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209‐249. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
