Human motor neurons derived from induced pluripotent stem cells are susceptible to SARS-CoV-2 infection
- PMID: 38116398
- PMCID: PMC10728732
- DOI: 10.3389/fncel.2023.1285836
Human motor neurons derived from induced pluripotent stem cells are susceptible to SARS-CoV-2 infection
Abstract
Introduction: COVID-19 typically causes Q7 respiratory disorders, but a high proportion of patients also reports neurological and neuromuscular symptoms during and after SARSCoV-2 infection. Despite a number of studies documenting SARS-CoV-2 infection of various neuronal cell populations, the impact of SARS-CoV-2 exposure on motor neuronal cells specifically has not been investigated so far.
Methods: Thus, by using human iPSC-derived motor neurons (iPSC-MNs) we assessed: (i) the expression of SARS-CoV-2 main receptors; (ii) iPSC-MN infectability by SARS-CoV-2; and (iii) the effect of SARS-CoV-2 exposure on iPSC-MN transcriptome.
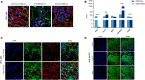
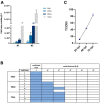
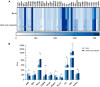
Results: Gene expression profiling and immunofluorescence (IF) analysis of the main host cell receptors recognized by SARS-CoV-2 revealed that all of them are expressed in iPSC-MNs, with CD147 and NRP1 being the most represented ones. By analyzing SARS-CoV-2 N1 and N2 gene expression over time, we observed that human iPSC-MNs were productively infected by SARS-CoV-2 in the absence of cytopathic effect. Supernatants collected from SARS-CoV-2-infected iPSC-MNs were able to re-infect VeroE6 cells. Image analyses of SARS-CoV-2 nucleocapsid proteins by IF confirmed iPSC-MN infectability. Furthermore, SARS-CoV-2 infection in iPSCMNs significantly altered the expression of genes (IL-6, ANG, S1PR1, BCL2, BAX, Casp8, HLA-A, ERAP1, CD147, MX1) associated with cell survival and metabolism, as well as antiviral and inflammatory response.
Discussion: These results suggest for the very first time that SARS-CoV-2 can productively infect human iPSC-derived MNs probably by binding CD147 and NRP1 receptors. Such information will be important to unveil the biological bases of neuromuscular disorders characterizing SARS-CoV-2 infection and the so called long-COVID symptoms.
Keywords: COVID-19; SARS-CoV-2 infection; iPSC-derived motor neurons; long-COVID; neuroinflammation; neuromuscular disorders.
Copyright © 2023 Cappelletti, Colombrita, Limanaqi, Invernizzi, Garziano, Vanetti, Moscheni, Santangelo, Zecchini, Trabattoni, Silani, Clerici, Ratti and Biasin.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures


Similar articles
-
BSG/CD147 and ACE2 receptors facilitate SARS-CoV-2 infection of human iPS cell-derived kidney podocytes.bioRxiv [Preprint]. 2021 Nov 17:2021.11.16.468893. doi: 10.1101/2021.11.16.468893. bioRxiv. 2021. Update in: Front Cell Dev Biol. 2022 Apr 20;10:855340. doi: 10.3389/fcell.2022.855340. PMID: 34816259 Free PMC article. Updated. Preprint.
-
SARS-CoV-2 Infection of Human Neurons Is TMPRSS2 Independent, Requires Endosomal Cell Entry, and Can Be Blocked by Inhibitors of Host Phosphoinositol-5 Kinase.J Virol. 2023 Apr 27;97(4):e0014423. doi: 10.1128/jvi.00144-23. Epub 2023 Apr 11. J Virol. 2023. PMID: 37039676 Free PMC article.
-
Human iPSC-Derived Cardiomyocytes Are Susceptible to SARS-CoV-2 Infection.Cell Rep Med. 2020 Jul 21;1(4):100052. doi: 10.1016/j.xcrm.2020.100052. Epub 2020 Jun 29. Cell Rep Med. 2020. PMID: 32835305 Free PMC article.
-
SARS-CoV-2 induces barrier damage and inflammatory responses in the human iPSC-derived intestinal epithelium.J Pharmacol Sci. 2022 Jul;149(3):139-146. doi: 10.1016/j.jphs.2022.04.010. Epub 2022 May 2. J Pharmacol Sci. 2022. PMID: 35641026 Free PMC article. Review.
-
Modeling Cardiac SARS-CoV-2 Infection with Human Pluripotent Stem Cells.Curr Cardiol Rep. 2022 Dec;24(12):2121-2129. doi: 10.1007/s11886-022-01813-2. Epub 2022 Oct 22. Curr Cardiol Rep. 2022. PMID: 36272051 Free PMC article. Review.
Cited by
-
Phase Separation of SARS-CoV-2 Nucleocapsid Protein with TDP-43 Is Dependent on C-Terminus Domains.Int J Mol Sci. 2024 Aug 12;25(16):8779. doi: 10.3390/ijms25168779. Int J Mol Sci. 2024. PMID: 39201466 Free PMC article.
-
Post COVID-19 Reflections and Questions: How Prepared Are We for the Next Pandemic?Int J Mol Sci. 2024 Jan 10;25(2):859. doi: 10.3390/ijms25020859. Int J Mol Sci. 2024. PMID: 38255933 Free PMC article.
-
Neuroimmune pathophysiology of long COVID.Psychiatry Clin Neurosci. 2025 Jun 19:10.1111/pcn.13855. doi: 10.1111/pcn.13855. Online ahead of print. Psychiatry Clin Neurosci. 2025. PMID: 40536011 Free PMC article.
-
Inflammatory Response and Defects on Myelin Integrity in the Olfactory System of K18hACE2 Mice Infected with SARS-CoV-2.eNeuro. 2024 Jun 17;11(6):ENEURO.0106-24.2024. doi: 10.1523/ENEURO.0106-24.2024. Print 2024 Jun. eNeuro. 2024. PMID: 38834299 Free PMC article.
-
SARS-CoV-2 natural infection, but not vaccine-induced immunity, elicits cross-reactive immunity to OC43.Heliyon. 2024 Sep 21;10(19):e37928. doi: 10.1016/j.heliyon.2024.e37928. eCollection 2024 Oct 15. Heliyon. 2024. PMID: 39391514 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

