SOCfinder: a genomic tool for identifying social genes in bacteria
- PMID: 38117204
- PMCID: PMC10763506
- DOI: 10.1099/mgen.0.001171
SOCfinder: a genomic tool for identifying social genes in bacteria
Abstract
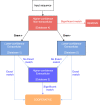
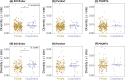
Bacteria cooperate by working collaboratively to defend their colonies, share nutrients, and resist antibiotics. Nevertheless, our understanding of these remarkable behaviours primarily comes from studying a few well-characterized species. Consequently, there is a significant gap in our understanding of microbial social traits, particularly in natural environments. To address this gap, we can use bioinformatic tools to identify genes that control cooperative or otherwise social traits. Existing tools address this challenge through two approaches. One approach is to identify genes that encode extracellular proteins, which can provide benefits to neighbouring cells. An alternative approach is to predict gene function using annotation tools. However, these tools have several limitations. Not all extracellular proteins are cooperative, and not all cooperative behaviours are controlled by extracellular proteins. Furthermore, existing functional annotation methods frequently miss known cooperative genes. We introduce SOCfinder as a new tool to find bacterial genes that control cooperative or otherwise social traits. SOCfinder combines information from several methods, considering if a gene is likely to [1] code for an extracellular protein [2], have a cooperative functional annotation, or [3] be part of the biosynthesis of a cooperative secondary metabolite. We use data on two extensively-studied species (P. aeruginosa and B. subtilis) to show that SOCfinder is better at finding known cooperative genes than existing tools. We also use theory from population genetics to identify a signature of kin selection in SOCfinder cooperative genes, which is lacking in genes identified by existing tools. SOCfinder opens up a number of exciting directions for future research, and is available to download from https://github.com/lauriebelch/SOCfinder.
Keywords: cooperation; cooperative gene; kin selection; relatedness; social gene.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

