Single-cell RNA-sequencing of PBMCs from SAVI patients reveals disease-associated monocytes with elevated integrated stress response
- PMID: 38118407
- PMCID: PMC10772457
- DOI: 10.1016/j.xcrm.2023.101333
Single-cell RNA-sequencing of PBMCs from SAVI patients reveals disease-associated monocytes with elevated integrated stress response
Abstract
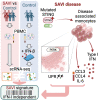
Gain-of-function mutations in stimulator of interferon gene 1 (STING1) result in STING-associated vasculopathy with onset in infancy (SAVI), a severe autoinflammatory disease. Although elevated type I interferon (IFN) production is thought to be the leading cause of the symptoms observed in patients, STING can induce a set of pathways, which have roles in the onset and severity of SAVI and remain to be elucidated. To this end, we performed a multi-omics comparative analysis of peripheral blood mononuclear cells (PBMCs) and plasma from SAVI patients and healthy controls, combined with a dataset of healthy PBMCs treated with IFN-β. Our data reveal a subset of disease-associated monocyte, expressing elevated CCL3, CCL4, and IL-6, as well as a strong integrated stress response, which we suggest is the result of direct PERK activation by STING. Cell-to-cell communication inference indicates that these monocytes lead to T cell early activation, resulting in their senescence and apoptosis. Last, we propose a transcriptomic signature of STING activation, independent of type I IFN response.
Keywords: SAVI; STING; integrated stress response; scRNA-seq; type I interferon; unfolded protein response.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests C.C., F.R.L., and M.M.M. are listed as inventors on a patent application related to this article (European Patent Application no. PCT/FR2023/050433, entitled “A gene signature for diagnosing stimulator of interferon genes (STING)-associated vasculopathy with onset in infancy (SAVI)”). F.R.L. and M.M.M. received grants from Sanofi (iAward Europe and research collaboration contract). C.C., L.D., M.D., F.A., G.B., J.C.G., and A.R. are or were employees of Sanofi and may hold shares and/or stock options in the company.
Figures






References
-
- Jeremiah N., Neven B., Gentili M., Callebaut I., Maschalidi S., Stolzenberg M.-C., Goudin N., Frémond M.L., Nitschke P., Molina T.J., et al. Inherited STING-activating mutation underlies a familial inflammatory syndrome with lupus-like manifestations. J. Clin. Invest. 2014;124:5516–5520. - PMC - PubMed
-
- Frémond M.L., Hadchouel A., Berteloot L., Melki I., Bresson V., Barnabei L., Jeremiah N., Belot A., Bondet V., Brocq O., et al. Overview of STING-Associated Vasculopathy with Onset in Infancy (SAVI) Among 21 Patients. J. Allergy Clin. Immunol. Pract. 2021;9:803–818.e11. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

