Pan-viral serology uncovers distinct virome patterns as risk predictors of hepatocellular carcinoma and intrahepatic cholangiocarcinoma
- PMID: 38118412
- PMCID: PMC10772458
- DOI: 10.1016/j.xcrm.2023.101328
Pan-viral serology uncovers distinct virome patterns as risk predictors of hepatocellular carcinoma and intrahepatic cholangiocarcinoma
Abstract
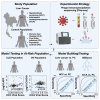
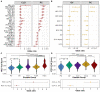
This study evaluates the pan-serological profiles of hepatocellular carcinoma (HCC) and intrahepatic cholangiocarcinoma (iCCA) compared to several diseased and non-diseased control populations to identify risk factors and biomarkers of liver cancer. We used phage immunoprecipitation sequencing, an anti-viral antibody screening method using a synthetic-phage-displayed human virome epitope library, to screen patient serum samples for exposure to over 1,280 strains of pathogenic and non-pathogenic viruses. Using machine learning methods to develop an HCC or iCCA viral score, we discovered that both viral scores were positively associated with several liver function markers in two separate at-risk populations independent of viral hepatitis status. The HCC score predicted all-cause mortality over 8 years in patients with chronic liver disease at risk of HCC, while the viral hepatitis status was not predictive of survival. These results suggest that non-hepatitis viral infections may contribute to HCC and iCCA development and could be biomarkers in at-risk populations.
Keywords: HCC; cholangiocarcinoma; hepatocellular carcinoma; iCCA; liver cancer; phage immunoprecipitation sequencing; phip-seq; serology; viral history; viruses.
Copyright © 2023. Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
-
- Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA. Cancer J. Clin. 2021;71:209–249. - PubMed
-
- Islami F., Goding Sauer A., Miller K.D., Siegel R.L., Fedewa S.A., Jacobs E.J., McCullough M.L., Patel A.V., Ma J., Soerjomataram I., et al. Proportion and number of cancer cases and deaths attributable to potentially modifiable risk factors in the United States. CA. Cancer J. Clin. 2018;68:31–54. - PubMed
-
- Kansagara D., Papak J., Pasha A.S., O’Neil M., Freeman M., Relevo R., Quiñones A., Motu’apuaka M., Jou J.H. Screening for Hepatocellular Carcinoma in Chronic Liver Disease. Ann. Intern. Med. 2014;161:261–269. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

