PRMT blockade induces defective DNA replication stress response and synergizes with PARP inhibition
- PMID: 38118413
- PMCID: PMC10772459
- DOI: 10.1016/j.xcrm.2023.101326
PRMT blockade induces defective DNA replication stress response and synergizes with PARP inhibition
Abstract
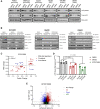
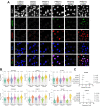
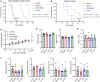
Multiple cancers exhibit aberrant protein arginine methylation by both type I arginine methyltransferases, predominately protein arginine methyltransferase 1 (PRMT1) and to a lesser extent PRMT4, and by type II PRMTs, predominately PRMT5. Here, we perform targeted proteomics following inhibition of PRMT1, PRMT4, and PRMT5 across 12 cancer cell lines. We find that inhibition of type I and II PRMTs suppresses phosphorylated and total ATR in cancer cells. Loss of ATR from PRMT inhibition results in defective DNA replication stress response activation, including from PARP inhibitors. Inhibition of type I and II PRMTs is synergistic with PARP inhibition regardless of homologous recombination function, but type I PRMT inhibition is more toxic to non-malignant cells. Finally, we demonstrate that the combination of PARP and PRMT5 inhibition improves survival in both BRCA-mutant and wild-type patient-derived xenografts without toxicity. Taken together, these results demonstrate that PRMT5 inhibition may be a well-tolerated approach to sensitize tumors to PARP inhibition.
Keywords: DNA replication stress; PARP inhibitors; PRMT inhibitors; arginine methylation; breast cancer; ovarian cancer.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests M.T.L. is a founder and limited partner in StemMed, Ltd., and a manager in StemMed Holdings, its general partner. He is a founder and equity stakeholder in Tvardi Therapeutics, Inc. Some PDXs are exclusively licensed to StemMed, Ltd., resulting in royalty income to M.T.L. L.E.D. is a compensated employee of StemMed, Ltd. Some PDXs are exclusively licensed to StemMed, Ltd., resulting in royalty income to L.E.D. A.K.S. discloses the following competing interests: consulting (Merck, GSK, ImmunoGen, Iylon, Kiyatec, Astra Zeneca, Onxeo), shareholder (BioPath), and patent (EGFL6 antibody).
Figures



References
-
- Fuentes-Antrás J., Alcaraz-Sanabria A.L., Morafraile E.C., Noblejas-López M.D.M., Galán-Moya E.M., Baliu-Pique M., López-Cade I., García-Barberán V., Pérez-Segura P., Manzano A., et al. Mapping of Genomic Vulnerabilities in the Post-Translational Ubiquitination, SUMOylation and Neddylation Machinery in Breast Cancer. Cancers. 2021;13 - PMC - PubMed
-
- Kumar M., Joshi G., Chatterjee J., Kumar R. Epidermal Growth Factor Receptor and its Trafficking Regulation by Acetylation: Implication in Resistance and Exploring the Newer Therapeutic Avenues in Cancer. Curr. Top. Med. Chem. 2020;20:1105–1123. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

