Accelerating multiplexed profiling of protein-ligand interactions: High-throughput plate-based reactive cysteine profiling with minimal input
- PMID: 38118439
- PMCID: PMC10960705
- DOI: 10.1016/j.chembiol.2023.11.015
Accelerating multiplexed profiling of protein-ligand interactions: High-throughput plate-based reactive cysteine profiling with minimal input
Abstract
Chemoproteomics has made significant progress in investigating small-molecule-protein interactions. However, the proteome-wide profiling of cysteine ligandability remains challenging to adapt for high-throughput applications, primarily due to a lack of platforms capable of achieving the desired depth using low input in 96- or 384-well plates. Here, we introduce a revamped, plate-based platform which enables routine interrogation of either ∼18,000 or ∼24,000 reactive cysteines based on starting amounts of 10 or 20 μg, respectively. This represents a 5-10X reduction in input and 2-3X improved coverage. We applied the platform to screen 192 electrophiles in the native HEK293T proteome, mapping the ligandability of 38,450 reactive cysteines from 8,274 human proteins. We further applied the platform to characterize new cellular targets of established drugs, uncovering that ARS-1620, a KRASG12C inhibitor, binds to and inhibits an off-target adenosine kinase ADK. The platform represents a major step forward to high-throughput proteome-wide evaluation of reactive cysteines.
Keywords: ABPP; TMT; chemoproteomics; covalent electrophile; cysteine; high throughput.
Copyright © 2023 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of interests S.P.G. is on the advisory board for Thermo Fisher Scientific, Cedilla Therapeutics, Casma Therapeutics, Cell Signaling Technology, and Frontier Medicines.
Figures


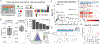


Comment in
-
High-throughput profiling of reactive cysteines.Nat Rev Drug Discov. 2024 Feb;23(2):107. doi: 10.1038/d41573-024-00008-4. Nat Rev Drug Discov. 2024. PMID: 38216770 No abstract available.
References
-
- Pace NJ, and Weerapana E (2013). Diverse functional roles of reactive cysteines. ACS Chem Biol 8, 283–296. - PubMed
-
- Janes MR, Zhang J, Li LS, Hansen R, Peters U, Guo X, Chen Y, Babbar A, Firdaus SJ, Darjania L, et al. (2018). Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor. Cell 172, 578–589 e517. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

