Development of a Rapid Adeno-Associated Virus (AAV) Identity Testing Platform through Comprehensive Intact Mass Analysis of Full-Length AAV Capsid Proteins
- PMID: 38123456
- PMCID: PMC10775144
- DOI: 10.1021/acs.jproteome.3c00513
Development of a Rapid Adeno-Associated Virus (AAV) Identity Testing Platform through Comprehensive Intact Mass Analysis of Full-Length AAV Capsid Proteins
Abstract
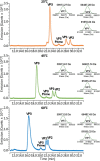
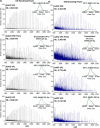
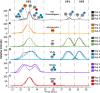
Adeno-associated viruses (AAVs) are commonly used as vectors for the delivery of gene therapy targets. Characterization of AAV capsid proteins (VPs) and their post-translational modifications (PTMs) have become a critical attribute monitored to evaluate product quality. Liquid chromatography-mass spectrometry (LC-MS) analysis of intact AAV VPs provides both quick and reliable serotype identification as well as proteoform information on each VP. Incorporating these analytical strategies into rapid good manufacturing practice (GMP)-compliant workflows containing robust, but simplified, data processing methods is necessary to ensure effective product quality control (QC) during production. Here, we present a GMP-compliant LC-MS workflow for the rapid identification and in-depth characterization of AAVs. Hydrophilic interaction liquid chromatography (HILIC) MS with difluoroacetic acid as a mobile phase modifier is utilized to achieve the intact separation and identification of AAV VPs and their potential proteoforms. Peptide mapping is performed to confirm PTMs identified during intact VP analysis and for in-depth PTM characterization. The intact separations platform is then incorporated into a data processing workflow developed using GMP-compliant software capable of rapid AAV serotype identification and, if desired, specific serotype PTM monitoring and characterization. Such a platform provides product QC capabilities that are easily accessible in a regulatory setting.
Keywords: AAV intact viral capsid protein screening; adeno-associated virus; cell and gene therapy; good manufacturing practices; hydrophilic interaction liquid chromatography−mass spectrometry; rapid identity testing.
Conflict of interest statement
The authors declare the following competing financial interest(s): S.G.M. and R.O.S. are employees of Patheon Viral Vector Services. J.B. received funding from Patheon Viral Vector Services to undertake this research. J.S. was employed under the collaborative research engagement between Patheon Viral Vector Services and NIBRT.
Figures


Similar articles
-
Characterization of Adeno-Associated Virus Capsid Proteins Using Hydrophilic Interaction Chromatography Coupled with Mass Spectrometry.J Pharm Biomed Anal. 2020 Sep 10;189:113481. doi: 10.1016/j.jpba.2020.113481. Epub 2020 Jul 21. J Pharm Biomed Anal. 2020. PMID: 32750536
-
Direct Liquid Chromatography/Mass Spectrometry Analysis for Complete Characterization of Recombinant Adeno-Associated Virus Capsid Proteins.Hum Gene Ther Methods. 2017 Oct;28(5):255-267. doi: 10.1089/hgtb.2016.178. Epub 2017 Jun 16. Hum Gene Ther Methods. 2017. PMID: 28627251
-
Optimized Reversed-Phase Liquid Chromatography/Mass Spectrometry Methods for Intact Protein Analysis and Peptide Mapping of Adeno-Associated Virus Proteins.Hum Gene Ther. 2021 Dec;32(23-24):1501-1511. doi: 10.1089/hum.2021.046. Epub 2021 Sep 20. Hum Gene Ther. 2021. PMID: 34278837 Free PMC article.
-
The role of the adeno-associated virus capsid in gene transfer.Methods Mol Biol. 2008;437:51-91. doi: 10.1007/978-1-59745-210-6_2. Methods Mol Biol. 2008. PMID: 18369962 Free PMC article. Review.
-
Recent Advances in the Analysis Full/Empty Capsid Ratio and Genome Integrity of Adeno-associated Virus (AAV) Gene Delivery Vectors.Curr Mol Med. 2020;20(10):806-813. doi: 10.2174/1566524020999200730181042. Curr Mol Med. 2020. PMID: 32748744 Review.
Cited by
-
A Direct Comparison of rAAV5 Variants Derived from the Baculovirus Expression System Using LC-MS Workflows Demonstrates Key Differences in Overall Production Yield, Product Quality and Vector Efficiency.Int J Mol Sci. 2024 Feb 28;25(5):2785. doi: 10.3390/ijms25052785. Int J Mol Sci. 2024. PMID: 38474031 Free PMC article.
-
Combination of hydrophilic interaction liquid chromatography and top-down mass spectrometry for characterisation of adeno-associated virus capsid proteins.Anal Bioanal Chem. 2025 Jun;417(15):3405-3417. doi: 10.1007/s00216-025-05874-4. Epub 2025 Apr 21. Anal Bioanal Chem. 2025. PMID: 40259015 Free PMC article.
-
Development of an HEK293 Suspension Cell Culture Medium, Transient Transfection Optimization Workflow, and Analytics for Batch rAAV Manufacturing.Biotechnol Bioeng. 2025 Jul;122(7):1640-1655. doi: 10.1002/bit.28980. Epub 2025 Apr 8. Biotechnol Bioeng. 2025. PMID: 40197832 Free PMC article.
-
Top-down mass spectrometry analysis of capsid proteins of recombinant adeno-associated virus using multiple ion activations and proton transfer charge reduction.Proteomics. 2025 Mar;25(5-6):e2400223. doi: 10.1002/pmic.202400223. Epub 2024 Sep 5. Proteomics. 2025. PMID: 39233542
References
LinkOut - more resources
Full Text Sources
Miscellaneous

