A unique sigma/anti-sigma system in the actinomycete Actinoplanes missouriensis
- PMID: 38123564
- PMCID: PMC10733313
- DOI: 10.1038/s41467-023-44291-y
A unique sigma/anti-sigma system in the actinomycete Actinoplanes missouriensis
Abstract
Bacteria of the genus Actinoplanes form sporangia that contain dormant sporangiospores which, upon contact with water, release motile spores (zoospores) through a process called sporangium dehiscence. Here, we set out to study the molecular mechanisms behind sporangium dehiscence in Actinoplanes missouriensis and discover a sigma/anti-sigma system with unique features. Protein σSsdA contains a functional sigma factor domain and an anti-sigma factor antagonist domain, while protein SipA contains an anti-sigma factor domain and an anti-sigma factor antagonist domain. Remarkably, the two proteins interact with each other via the anti-sigma factor antagonist domain of σSsdA and the anti-sigma factor domain of SipA. Although it remains unclear whether the SipA/σSsdA system plays direct roles in sporangium dehiscence, the system seems to modulate oxidative stress responses in zoospores. In addition, we identify a two-component regulatory system (RsdK-RsdR) that represses initiation of sporangium dehiscence.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



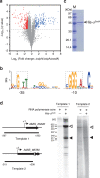
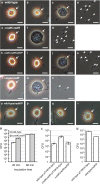
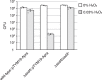
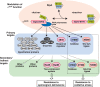
References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- JP26252010/Ministry of Education, Culture, Sports, Science and Technology (MEXT)
- JP18H02122/Ministry of Education, Culture, Sports, Science and Technology (MEXT)
- JP19H05685/Ministry of Education, Culture, Sports, Science and Technology (MEXT)
- A3 Foresight Program/MEXT | Japan Society for the Promotion of Science (JSPS)
- JP16H06279/MEXT | Japan Society for the Promotion of Science (JSPS)
LinkOut - more resources
Full Text Sources

