Single-cell multi-omics analysis of human testicular germ cell tumor reveals its molecular features and microenvironment
- PMID: 38123589
- PMCID: PMC10733385
- DOI: 10.1038/s41467-023-44305-9
Single-cell multi-omics analysis of human testicular germ cell tumor reveals its molecular features and microenvironment
Abstract
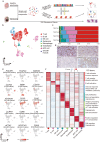
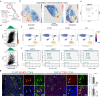
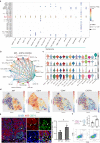
Seminoma is the most common malignant solid tumor in 14 to 44 year-old men. However, its molecular features and tumor microenvironment (TME) is largely unexplored. Here, we perform a series of studies via genomics profiling (single cell multi-omics and spatial transcriptomics) and functional examination using seminoma samples and a seminoma cell line. We identify key gene expression programs share between seminoma and primordial germ cells, and further characterize the functions of TFAP2C in promoting tumor invasion and migration. We also identify 15 immune cell subtypes in TME, and find that subtypes with exhaustion features were located closer to the tumor region through combined spatial transcriptome analysis. Furthermore, we identify key pathways and genes that may facilitate seminoma disseminating beyond the seminiferous tubules. These findings advance our knowledge of seminoma tumorigenesis and produce a multi-omics atlas of in situ human seminoma microenvironment, which could help discover potential therapy targets for seminoma.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

