The PfRCR complex bridges malaria parasite and erythrocyte during invasion
- PMID: 38123677
- PMCID: PMC10794152
- DOI: 10.1038/s41586-023-06856-1
The PfRCR complex bridges malaria parasite and erythrocyte during invasion
Abstract
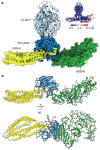
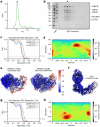
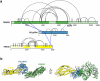
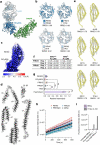
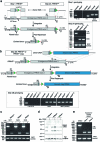
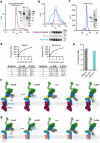
The symptoms of malaria occur during the blood stage of infection, when parasites invade and replicate within human erythrocytes. The PfPCRCR complex1, containing PfRH5 (refs. 2,3), PfCyRPA, PfRIPR, PfCSS and PfPTRAMP, is essential for erythrocyte invasion by the deadliest human malaria parasite, Plasmodium falciparum. Invasion can be prevented by antibodies3-6 or nanobodies1 against each of these conserved proteins, making them the leading blood-stage malaria vaccine candidates. However, little is known about how PfPCRCR functions during invasion. Here we present the structure of the PfRCR complex7,8, containing PfRH5, PfCyRPA and PfRIPR, determined by cryogenic-electron microscopy. We test the hypothesis that PfRH5 opens to insert into the membrane9, instead showing that a rigid, disulfide-locked PfRH5 can mediate efficient erythrocyte invasion. We show, through modelling and an erythrocyte-binding assay, that PfCyRPA-binding antibodies5 neutralize invasion through a steric mechanism. We determine the structure of PfRIPR, showing that it consists of an ordered, multidomain core flexibly linked to an elongated tail. We also show that the elongated tail of PfRIPR, which is the target of growth-neutralizing antibodies6, binds to the PfCSS-PfPTRAMP complex on the parasite membrane. A modular PfRIPR is therefore linked to the merozoite membrane through an elongated tail, and its structured core presents PfCyRPA and PfRH5 to interact with erythrocyte receptors. This provides fresh insight into the molecular mechanism of erythrocyte invasion and opens the way to new approaches in rational vaccine design.
© 2023. The Author(s).
Conflict of interest statement
M.K.H. and S.J.D. are named inventors on patents related to the use of PfRH5 as a vaccine (no. GB2105/052205), to the design of a thermally stabilized version of PfRH5 for use as a vaccine (no. GB2017/052608) and to mABs targeting PfRH5 (no. GB2019/052885). B.F., N.A., M.N.H., A.J., R.J.R., H.W.-M. and E.K. declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

