Emergence of replication timing during early mammalian development
- PMID: 38123678
- PMCID: PMC10781638
- DOI: 10.1038/s41586-023-06872-1
Emergence of replication timing during early mammalian development
Abstract
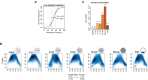
DNA replication enables genetic inheritance across the kingdoms of life. Replication occurs with a defined temporal order known as the replication timing (RT) programme, leading to organization of the genome into early- or late-replicating regions. RT is cell-type specific, is tightly linked to the three-dimensional nuclear organization of the genome1,2 and is considered an epigenetic fingerprint3. In spite of its importance in maintaining the epigenome4, the developmental regulation of RT in mammals in vivo has not been explored. Here, using single-cell Repli-seq5, we generated genome-wide RT maps of mouse embryos from the zygote to the blastocyst stage. Our data show that RT is initially not well defined but becomes defined progressively from the 4-cell stage, coinciding with strengthening of the A and B compartments. We show that transcription contributes to the precision of the RT programme and that the difference in RT between the A and B compartments depends on RNA polymerase II at zygotic genome activation. Our data indicate that the establishment of nuclear organization precedes the acquisition of defined RT features and primes the partitioning of the genome into early- and late-replicating domains. Our work sheds light on the establishment of the epigenome at the beginning of mammalian development and reveals the organizing principles of genome organization.
© 2023. The Author(s).
Conflict of interest statement
M.-E.T.-P. is a member of the ethics advisory panel of MERCK. The other authors declare no competing interests.
Figures


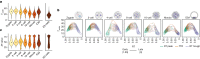











References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

