Fluorescence Correlation Spectroscopy as a Versatile Method to Define Aptamer-Protein Interactions with Single-Molecule Sensitivity
- PMID: 38124657
- PMCID: PMC10782416
- DOI: 10.1021/acs.analchem.3c03341
Fluorescence Correlation Spectroscopy as a Versatile Method to Define Aptamer-Protein Interactions with Single-Molecule Sensitivity
Abstract
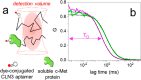
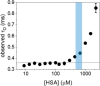
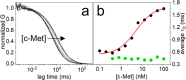
Aptamers are folded oligonucleotides that selectively recognize and bind a target and are consequently regarded as an emerging alternative to antibodies for sensing and therapeutic applications. The rational development of functional aptamers is strictly related to the accurate definition of molecular binding properties. Nevertheless, most of the methodologies employed to define binding affinities use bulk measurements. Here, we describe the use of fluorescence correlation spectroscopy (FCS) as a method with single-molecule sensitivity that quantitatively defines aptamer-protein binding. First, FCS was used to measure the equilibrium affinity between the CLN3 aptamer, conjugated with a dye, and its target, the c-Met protein. Equilibrium affinity was also determined for other functional aptamers targeting nucleolin and platelet-derived growth factors. Then, association and dissociation rates of CLN3 to/from the target protein were measured using FCS by monitoring the equilibration kinetics of the binding reaction in solution. Finally, FCS was exploited to investigate the behavior of CLN3 exposed to physiological concentrations of the most abundant serum proteins. Under these conditions, the aptamer showed negligible interactions with nontarget serum proteins while preserving its affinity for the c-Met. The presented results introduce FCS as an alternative or complementary analytical tool in aptamer research, particularly well-suited for the characterization of protein-targeting aptamers.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

