A SARS-CoV-2 NSP7 homolog of a Treg epitope suppresses CD4+ and CD8+ T cell memory responses
- PMID: 38124752
- PMCID: PMC10731459
- DOI: 10.3389/fimmu.2023.1290688
A SARS-CoV-2 NSP7 homolog of a Treg epitope suppresses CD4+ and CD8+ T cell memory responses
Abstract
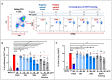
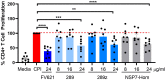
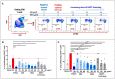
Pathogens escape host defenses by T-cell epitope mutation or deletion (immune escape) and by simulating the appearance of human T cell epitopes (immune camouflage). We identified a highly conserved, human-like T cell epitope in non-structural protein 7 (NSP7) of SARS-CoV-2, RNA-dependent RNA polymerase (RdRp) hetero-tetramer complex. Remarkably, this T cell epitope has significant homology to a T regulatory cell epitope (Tregitope) previously identified in the Fc region of human immunoglobulin G (IgG) (Tregitope 289). We hypothesized that the SARS-CoV-2 NSP7 epitope (NSP7-289) may induce suppressive responses by engaging and activating pre-existing regulatory T cells. We therefore compared NSP7-289 and IgG Tregitopes (289 and 289z, a shorter version of 289 that isolates the shared NSP7 epitope) in vitro. Tregitope peptides 289, 289z and NSP7-289 bound to multiple HLA-DRB1 alleles in vitro and suppressed CD4+ and CD8+ T cell memory responses. Identification and in vitro validation of SARS-CoV-2 NSP7-289 provides further evidence of immune camouflage and suggests that pathogens can use human-like epitopes to evade immune response and potentially enhance host tolerance. Further exploration of the role of cross-conserved Tregs in human immune responses to pathogens such as SARS-CoV-2 is warranted.
Keywords: SARS-CoV-2; Tregitope; immune camouflage; immunogenicity; nsp7; tolerance.
Copyright © 2023 Miah, Lelias, Gutierrez, McAllister, Boyle, Moise and De Groot.
Conflict of interest statement
ADG is the senior officer and shareholder of, and SM, SL, MM, CB and AHG are employees of EpiVax, Inc., a privately-owned biotechnology company located in Providence, RI. LM is a former employee of EpiVax, Inc. All of these authors acknowledge that there is a potential conflict of interest related to their employment status and/or ownership relationship with EpiVax and attest that the work contained in this research report is free of any bias that might be associated with the commercial goals of the company. This study received funding from EpiVax. The funder had the following involvement with the study: All in vitro and bioinformatics studies were performed after group discussion and decision making by the authors, and funded by the internal research funds of the EpiVax company.
Figures



References
-
- Worldometer.info . Worldometer COVID-19 data (2023). Available at: https://www.worldometers.info/coronavirus/.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

