Emerging evidence for dysregulated proteome cargoes of tau-propagating extracellular vesicles driven by familial mutations of tau and presenilin
- PMID: 38125374
- PMCID: PMC10732590
- DOI: 10.20517/evcna.2023.44
Emerging evidence for dysregulated proteome cargoes of tau-propagating extracellular vesicles driven by familial mutations of tau and presenilin
Abstract
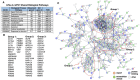
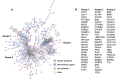
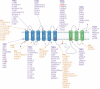
Tau propagation, pathogenesis, and neurotoxicity are hallmarks of neurodegenerative diseases that result in cognitive impairment. Tau accumulates in Alzheimer's disease (AD), frontotemporal dementia and parkinsonism linked to chromosome 17 (FTDP-17), chronic traumatic encephalopathy (CTE), progressive supranuclear palsy, and related tauopathies. Knowledge of the mechanisms for tau propagation in neurodegeneration is necessary for understanding the development of dementia. Exosomes, known as extracellular vesicles (EVs), have emerged as participants in promoting tau propagation. Recent findings show that EVs generated by neurons expressing familial mutations of tauopathies of FTDP-17 (P301L and V337M) (mTau) and presenilin (A246E) (mPS1) in AD induce tau propagation and accumulation after injection into rodent brain. To gain knowledge of the proteome cargoes of the mTau and mPS1 EVs that promote tau pathogenesis, this review compares the proteomes of these EVs, which results in important new questions concerning EV mechanisms of tau pathogenesis. Proteomics data show that EVs produced by mTau- and mPS1-expressing iPSC neurons share proteins involved in exocytosis and vesicle secretion and, notably, these EVs also possess differences in protein components of vesicle-mediated transport, extracellular functions, and cell adhesion. It will be important for future studies to gain an understanding of the breadth of familial genetic mutations of tau, presenilin, and other genes in promoting EV initiation of tau propagation and pathogenesis. Furthermore, elucidation of EV cargo components that mediate tau propagation will have potential as biomarkers and therapeutic strategies to ameliorate dementia of tauopathies.
Keywords: Alzheimer’s disease; Tauopathies; exosome; extracellular vesicle; frontotemporal dementia; mutant presenilin; mutant tau; proteomics.
Conflict of interest statement
Conflicts of interest The authors declared that there are no conflicts of interest.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
