WASF3 overexpression affects the expression of circular RNA hsa-circ-0100153, which promotes breast cancer progression by sponging hsa-miR-31, hsa-miR-767-3p, and hsa-miR-935
- PMID: 38125536
- PMCID: PMC10731075
- DOI: 10.1016/j.heliyon.2023.e22874
WASF3 overexpression affects the expression of circular RNA hsa-circ-0100153, which promotes breast cancer progression by sponging hsa-miR-31, hsa-miR-767-3p, and hsa-miR-935
Abstract
Background: The WASF3 gene has been linked to promoting metastasis in breast cancer (BC) cells, and low expression reduces invasion potential. Circular RNAs (circRNAs) function as microRNA (miRNA) modulators and are involved in cancer progression, but the relationship between these factors remains unclear.
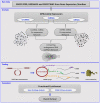
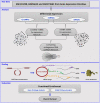
Methods: This study used bioinformatics methods and a computational approach to investigate the role of circRNAs and miRNAs in the context of WASF3 overexpression. Differentially expressed mRNAs, circRNAs, and miRNAs were identified using Gene Expression Omnibus (GEO) datasets. A competing endogenous RNA (ceRNA) network was constructed based on circRNA-miRNA pairs and miRNA-mRNA pairs. Functional and pathway enrichment analyses were predicted using a circRNA-miRNA-mRNA network.
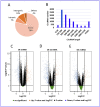
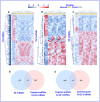
Results: RNA expression patterns were significantly different between normal and tumor samples. A total of 190 circRNAs, 76 miRNAs, and 678 mRNAs were differentially expressed. The analysis of the circRNA-miRNA-mRNA regulatory network revealed interactions between hsa-circ-0100153, hsa-miR-31, hsa-miR-767-3p, and hsa-miR-935 with WASF3 in cancer. These interactions primarily function in DNA replication and the cell cycle.
Conclusions: This study reveals a mechanism by which WASF3 overexpression affects the expression of circRNAs hsa-circ-0100153, promoting BC progression by sponging hsa-miR-31/hsa-miR-767-3p /hsa-miR-935. This mechanism may increase the invasive potential of cancers, in addition to other reported molecular mechanisms involving the WASF3 gene.
Keywords: Bioinformatics; Cancer; Gene expression; Oncology; circRNA; miRNA.
© 2023 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

Similar articles
-
Identification of the hsa_circ_0039466/miR-96-5p/FOXO1 regulatory network in hepatocellular carcinoma by whole-transcriptome analysis.Ann Transl Med. 2022 Jul;10(14):769. doi: 10.21037/atm-22-3147. Ann Transl Med. 2022. PMID: 35965793 Free PMC article.
-
Construction of a circRNA-miRNA-mRNA Regulatory Network for Coronary Artery Disease by Bioinformatics Analysis.Cardiol Res Pract. 2022 Feb 16;2022:4017082. doi: 10.1155/2022/4017082. eCollection 2022. Cardiol Res Pract. 2022. PMID: 35223093 Free PMC article.
-
The investigation of circRNA profiling reveals the regulatory role of the hsa_circ_0000375/miR-7706 pathway in breast cancer.Mol Biol Rep. 2023 Dec;50(12):9993-10004. doi: 10.1007/s11033-023-08798-3. Epub 2023 Oct 30. Mol Biol Rep. 2023. PMID: 37904009
-
Bioinformatics Analysis Predicts hsa_circ_0026337/miR-197-3p as a Potential Oncogenic ceRNA Network for Non-Small Cell Lung Cancers.Anticancer Agents Med Chem. 2022;22(5):874-886. doi: 10.2174/1871520621666210712090721. Anticancer Agents Med Chem. 2022. PMID: 34254931
-
Whole-transcriptome analysis reveals a potential hsa_circ_0001955/hsa_circ_0000977-mediated miRNA-mRNA regulatory sub-network in colorectal cancer.Aging (Albany NY). 2020 Mar 28;12(6):5259-5279. doi: 10.18632/aging.102945. Epub 2020 Mar 28. Aging (Albany NY). 2020. PMID: 32221048 Free PMC article.
Cited by
-
The endogenous association among MMP2/miR-1248/Circ_0087558/miR-643/ MAP2K6 axis can contribute to brain metastasis in basal-like subtype of breast cancer.Heliyon. 2024 Jun 24;10(13):e33195. doi: 10.1016/j.heliyon.2024.e33195. eCollection 2024 Jul 15. Heliyon. 2024. PMID: 39027611 Free PMC article.
-
Environmental cadmium-induced circRNA-miRNA-mRNA network regulatory mechanism in human normal liver cell model.Curr Res Toxicol. 2024 Aug 24;7:100192. doi: 10.1016/j.crtox.2024.100192. eCollection 2024. Curr Res Toxicol. 2024. PMID: 39280980 Free PMC article.
-
Unraveling non-coding RNAs in breast cancer: mechanistic insights and therapeutic potential.Med Oncol. 2024 Dec 27;42(1):37. doi: 10.1007/s12032-024-02589-x. Med Oncol. 2024. PMID: 39730979 Review.
-
Investigating the interplay between the mir-183/182/96 cluster and the adherens junction pathway in early-stage breast cancer.Sci Rep. 2024 Oct 21;14(1):24711. doi: 10.1038/s41598-024-73632-0. Sci Rep. 2024. PMID: 39433788 Free PMC article.
References
-
- Ghufran MdS., Soni P., Duddukuri G.R. In: Bioprospecting Trop Med Plants [Internet] Arunachalam K., Yang X., Puthanpura Sasidharan S., editors. Springer Nature Switzerland; Cham: 2023. The global concern for cancer emergence and its prevention: a systematic unveiling of the present scenario. [cited 2023 Nov 1]. pp. 1429–1455. Available from: - DOI
-
- Henley S.J., Ward E.M., Scott S., Ma J., Anderson R.N., Firth A.U., Thomas C.C., Islami F., Weir H.K., Lewis D.R., Sherman R.L., Wu M., Benard V.B., Richardson L.C., Jemal A., Cronin K., Kohler B.A. Annual report to the nation on the status of cancer, part I: national cancer statistics. Cancer. 2020 May 15;126(10):2225–2249. PMCID: PMC7299151. - PMC - PubMed
LinkOut - more resources
Full Text Sources

