Gene dosage adaptations to mtDNA depletion and mitochondrial protein stress in budding yeast
- PMID: 38126114
- PMCID: PMC10849340
- DOI: 10.1093/g3journal/jkad272
Gene dosage adaptations to mtDNA depletion and mitochondrial protein stress in budding yeast
Abstract
Mitochondria contain a local genome (mtDNA) comprising a small number of genes necessary for respiration, mitochondrial transcription and translation, and other vital functions. Various stressors can destabilize mtDNA leading to mtDNA loss. While some cells can survive mtDNA loss, they exhibit various deficiencies. Here, we investigated the impact of proteotoxicity on mitochondrial function by inducing mitochondrial unfolded protein stress in budding yeast. This led to rapid mtDNA loss, but aerobic conditioning imparted transient resistance to mitochondrial protein stress. We present a quantitative model of mtDNA loss in a growing cell population and measure its parameters. To identify genetic adaptations to mtDNA depletion, we performed a genome-wide screen for gene dosage increases that affect the growth of cells lacking mtDNA. The screen revealed a set of dosage suppressors that alleviate the growth impairment in mtDNA-deficient cells. Additionally, we show that these suppressors of mtDNA stress both bolster cell proliferation and prevent mtDNA loss during mitochondrial protein stress.
Keywords: copy number variation; functional genomics; mitochondria; mtDNA; protein aggregates; proteostasis; respiration; stress resistance.
© The Author(s) 2023. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest The author(s) declare no conflicts of interest.
Figures
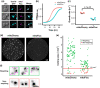

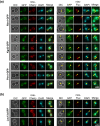

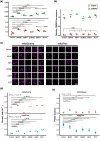
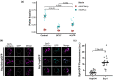
Similar articles
-
Global genetic determinants of mitochondrial DNA copy number.PLoS One. 2014 Aug 29;9(8):e105242. doi: 10.1371/journal.pone.0105242. eCollection 2014. PLoS One. 2014. PMID: 25170845 Free PMC article.
-
Mitochondrial fusion increases the mitochondrial DNA copy number in budding yeast.Genes Cells. 2011 May;16(5):527-44. doi: 10.1111/j.1365-2443.2011.01504.x. Epub 2011 Apr 5. Genes Cells. 2011. PMID: 21463454
-
Mitochondrial transcription factor A regulates mitochondrial transcription initiation, DNA packaging, and genome copy number.Biochim Biophys Acta. 2012 Sep-Oct;1819(9-10):921-9. doi: 10.1016/j.bbagrm.2012.03.002. Epub 2012 Mar 21. Biochim Biophys Acta. 2012. PMID: 22465614 Review.
-
Overexpression of TFAM or twinkle increases mtDNA copy number and facilitates cardioprotection associated with limited mitochondrial oxidative stress.PLoS One. 2015 Mar 30;10(3):e0119687. doi: 10.1371/journal.pone.0119687. eCollection 2015. PLoS One. 2015. PMID: 25822152 Free PMC article.
-
Number matters: control of mammalian mitochondrial DNA copy number.J Genet Genomics. 2009 Mar;36(3):125-31. doi: 10.1016/S1673-8527(08)60099-5. J Genet Genomics. 2009. PMID: 19302968 Free PMC article. Review.
Cited by
-
The micromammals.G3 (Bethesda). 2024 Jun 5;14(6):jkae073. doi: 10.1093/g3journal/jkae073. G3 (Bethesda). 2024. PMID: 38837137 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
