Whole-Blood Transcriptome Unveils Altered Immune Response in Acute Myocardial Infarction Patients With Aortic Valve Sclerosis
- PMID: 38126173
- PMCID: PMC10805353
- DOI: 10.1161/ATVBAHA.123.320106
Whole-Blood Transcriptome Unveils Altered Immune Response in Acute Myocardial Infarction Patients With Aortic Valve Sclerosis
Abstract
Background: Aortic valve sclerosis (AVSc) presents similar pathogenetic mechanisms to coronary artery disease and is associated with short- and long-term mortality in patients with coronary artery disease. Evidence of AVSc-specific pathophysiological traits in acute myocardial infarction (AMI) is currently lacking. Thus, we aimed to identify a blood-based transcriptional signature that could differentiate AVSc from no-AVSc patients during AMI.
Methods: Whole-blood transcriptome of AVSc (n=44) and no-AVSc (n=66) patients with AMI was assessed by RNA sequencing on hospital admission. Feature selection, differential expression, and enrichment analyses were performed to identify gene expression patterns discriminating AVSc from no-AVSc and infer functional associations. Multivariable Cox regression analysis was used to estimate the hazard ratios of cardiovascular events in AVSc versus no-AVSc patients.
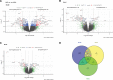
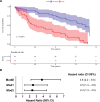
Results: This cross-sectional study identified a panel of 100 informative genes capable of distinguishing AVSc from no-AVSc patients with 94% accuracy. Further analysis revealed significant mean differences in 143 genes, of which 30 genes withstood correction for age and previous AMI or coronary interventions. Functional inference unveiled a significant association between AVSc and key biological processes, including acute inflammatory responses, type I IFN (interferon) response, platelet activation, and hemostasis. Notably, patients with AMI with AVSc exhibited a significantly higher incidence of adverse cardiovascular events during a 10-year follow-up period, with a full adjusted hazard ratio of 2.4 (95% CI, 1.3-4.5).
Conclusions: Our findings shed light on the molecular mechanisms underlying AVSc and provide potential prognostic insights for patients with AMI with AVSc. During AMI, patients with AVSc showed increased type I IFN (interferon) response and earlier adverse cardiovascular outcomes. Novel pharmacological therapies aiming at limiting type I IFN response during or immediately after AMI might improve poor cardiovascular outcomes of patients with AMI with AVSc.
Keywords: acute myocardial infarction; aortic valve sclerosis; cardiovascular adverse events; immune response; inflammation.
Conflict of interest statement
Figures




References
-
- Gharacholou SM, Karon BL, Shub C, Pellikka PA. Aortic valve sclerosis and clinical outcomes: moving toward a definition. Am J Med. 2011;124:103–110. doi: 10.1016/j.amjmed.2010.10.012 - PubMed
-
- Poggio P, Cavallotti L, Myasoedova VA, Bonomi A, Songia P, Gripari P, Valerio V, Amato M, Barbieri S, Faggiano P, et al. . Aortic valve sclerosis adds to prediction of short-term mortality in patients with documented coronary atherosclerosis. J Clin Med. 2019;8:1172. doi: 10.3390/jcm8081172 - PMC - PubMed
-
- Vasan RS. Biomarkers of cardiovascular disease: molecular basis and practical considerations. Circulation. 2006;113:2335–2362. doi: 10.1161/CIRCULATIONAHA.104.482570 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

